Abstract
We recently developed an original HCV-permissive cell culture model based on both partial differentiation and physiological oxygen pressure (Hypo-Diff cells) that produces highly infectious lipid-rich and native-like HCV-lipoviroparticles (LVPs). Here, we explored the precise role of physiological hypoxia and related specific transcription factors, hypoxia-inducible factors 1α and 2α (HIF-1α and HIF-2α) to better understand the mechanism governing viral morphogenesis. Knocking-down HIF-2α specifically reduced both the number and size of neutral lipid-rich droplets in Huh7.5 Hypo-Diff cells, suggesting a central role for HIF-2 in controlling lipid metabolism under physiological hypoxia. In HCV-infected siHIF-2α Hypo-Diff cells, both HCV replication and the specific infectious viral efficacy of progeny viruses were significantly impaired. Interestingly, the ectopic expression of a mutated form of the HIF-2α protein, stabilized in normoxia, both increased the number and size of neutral lipid-rich droplets and restored the production of highly infectious HCV viruses in the absence of cell differentiation. Finally, by iodixanol fractionation of supernatants to determine the mean buoyant density of infectious HCV LVPs, we established that HIF-2α is exclusively responsible for producing highly lipidated and broadly infectious HCV-LVPs by Hypo-Diff cells. These findings thus clearly establish the central role of physiological hypoxia, and notably HIF-2, in the production of highly infectious lipid-rich native-like HCV particles. Since physiological hypoxia is a shared characteristic in mammalian tissues, we propose to reconsider the role of natural oxygen tension and especially the role of HIF-2 in the life cycle of other lipid-associated viruses, whether hepatotropic or not.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00018-025-05739-0.
Keywords: Cell culture model, Hypoxia, HIF-2, Lipid droplets, HCV, Morphogenesis and infectivity
Introduction
Hepatitis C treatment with direct-acting antivirals (DAAs) has revolutionized therapeutic protocols that can nowadays cure over 95% of the treated patients, leading the WHO to set the goal of eliminating hepatitis C as a major public health issue by 2030 (WHO Global Hepatitis Report, 2024) [1]. However, to achieve this goal, several major barriers to HCV eradication remain, including insufficient screening of subjects for treatment, the lack of DAA prescription in many countries, together with emerging DAA's treatment failures in unusual, underestimated HCV subtypes [1–3]. In this context, both fundamental and clinical research efforts remain necessary to achieve the WHO’s goals.
Among the challenging fundamental HCV research to be addressed, deciphering accurately the natural intracellular HCV cycle is of major importance. The infectious cell culture model for HCV (HCVcc – JFH1 isolate) can reproduce this cycle from viral entry to extracellular production of progeny viral particles, in highly permissive subclones of human de-differentiated hepatoma-derived Huh7 cells [4, 5]. Viral assembly and morphogenesis are however poorly characterized. These steps have been reported to be closely related to cellular lipid metabolism, a feature that the model lacks unlike the end-stage differentiated primary human hepatocytes [6, 7]. Native HCV is actually unique in its ability to associate closely with lipoprotein components, forming hybrid particles known as lipoviroparticles (LVPs) [8–10]. On the one hand the infectivity of HCVcc LVPs produced in standard culture conditions is mostly associated with high density, low-lipidated particles while infectious LVPs found in the plasma of infected individuals are large, highly-lipidated and have low-density [6, 7, 11].
We have recently developed a HCVcc cell culture model combining partially differentiated Huh7.5 cells with DMSO and prolonged exposure to a physiological oxygen tension named hypoxia [12]. These cell culture conditions restore lipid metabolism, favoring completion of the viral cycle and the production of highly infectious and lipid-enriched LVPs. The viral particles also present a global ultrastructure and an envelope glycoprotein E1E2-complex conformation close to the ones circulating in infected individuals, making this model suitable for both the analysis of the last steps of the viral cycle and cellular factors involved.
The central transcription factors mediating the cellular response to low oxygen concentration are Hypoxia-inducible factor (HIFs), which are composed of two subunits—HIFα (HIF-1α, HIF-2α and HIF-3α) and HIFβ (or ANRT)—considering HIF-1α and HIF-2α as the best characterized HIF-α subunits. Under normal oxygen conditions (normoxia; 21%, vol/vol), HIF-α subunits are quickly degraded by the proteasome after hydroxylation by prolyl hydroxylase domain (PHD) enzymes of amino acids (including P402, P564 in HIF-1α and P405, P531 in HIF-2α), von Hippel-Lindau (VHL) protein binding, and ubiquitinylation (reviewed in [13] and [14]). In hypoxia, PHD modifying HIF-α are nonfunctional, HIF-α are stabilized and translocated into the nucleus, where they interact with the constitutively expressed HIFβ subunit. This transcriptional complex cooperates with the hypoxia response element (HRE) on promoters of genes involved in multiple pathways including lipid metabolism [15–17]. Here we hypothesized that hypoxia regulates the production of highly infectious and lipid-enriched LVPs in partially differentiated Huh7.5 cells cultured in a physiological hypoxia and aimed to determine the role of HIFs in this feature.
By both RNA silencing and HIFs-α trans-complementation approaches, we describe that HIF-2 is the major factor that triggers Huh7.5 lipid metabolism and controls both HCV replication and the production of lipid-enriched and highly infectious progeny LVPs in our native-like HCV culture model.
Materials and methods
Cells and treatments
Huh7.5 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 4.5 g/L D-glucose, 4 mM L-glutamine (Invitrogen), and supplemented with 100 U/mL penicillin, 100 µg/mL streptomycin (Invitrogen) and 10% fetal calf serum (Thermo Fisher Scientific). Standard de-differentiated Huh7.5 cells were cultured at atmospheric oxygen tension (Std—normoxia—21% O2 [vol/vol], 5% CO2 [vol/vol], 74% N2 [vol/vol]). These cells were used to seed 12-well plates at a density of 4 × 105 cells/well 24 h before use. Differentiated Huh7.5 cells cultured in hypoxia (Hypo-Diff) were seeded in 12-well plates coated with rat type I collagen (Sigma-Aldrich), at a density of 8 × 104 cells/well. At 40% confluence, the cell culture medium was replaced with complete DMEM supplemented with 1% DMSO (vol/vol). Cells were cultured for 15 days, during which time the medium was replaced with DMEM supplemented with 1% DMSO every two days. Nine days after DMSO exposure, Huh7.5 cells were transferred into a hypoxia workstation (HypoxyLab—Oxford Optronix, Abingdon, UK) and cultured for another six days at low oxygen tension (1% [vol/vol] O2, 5% [vol/vol] CO2, 94% [vol/vol] N2).
RNA Interference
Four days post transfer into the hypoxia workstation, Hypo-Diff Huh7.5 cells were transfected with 10 nM of small interfering RNAs (siRNAs) targeting HIF-1α (siHIF-1α—Assay ID: s6541; Ambion), HIF-2α (siHIF-2α Assay ID: s4699; Ambion) or a non-targeting control siRNA (All Stars negative control siRNA, Qiagen) using Lipofectamine RNAiMax (Invitrogen). The medium was replaced by complete DMEM supplemented with 1% DMSO 6 h post-transfection according to the manufacturer’s instructions.
Plasmids and transfection
HA-HIF2alpha-P405 A/P531 A-pcDNA3 and HA-HIF1alpha P402 A/P564 A-pcDNA3 (named thereafter HIF-1α mut and HIF-2α mut), a gift from William Kaelin (Addgene plasmid # 18,956; http://n2t.net/addgene:18956; RRID:Addgene_18956 and Addgene plasmid # 18955; http://n2t.net/addgene:18955; RRID:Addgene_18955, respectively), are CMV promoter-driven pcDNA3 plasmids expressing stabilized HIF-1α and HIF-2α [18]. These plasmids included 2 mutated amino acids, which were then no longer hydroxylated by the PHD. The mutant proteins are O2 insensitive and thus resistant to proteasome-specific degradation under normoxic conditions. Standard de-differentiated Huh7.5 cells cultivated in normoxia were used to seed a 12-well plate at a density of 1 × 105 cells/well. Transfections were performed with 1.5 µg of HIF-1α mut, HIF-2α mut or pcDNA3 plasmids using JetPEI (Life Technologies) as transfectant, according to the manufacturer’s instructions.
HCVcc production and infection
For HCVcc production, Huh7.5 cells were transfected with JFH1 RNA by electroporation (single pulse of 900 µF and 270 V), as previously described [5, 19–21]. Supernatants were collected 48 and 72 h post-transfection, passed through a filter with 0.45 µm pores, and stored at −80 °C. JFH1-transfected Huh7.5 cell supernatants were harvested after three post-transfection passages, to obtain high HCVcc titers. HCVcc were titrated as previously described [4].
Except otherwise stated, HCVcc infection experiments were performed for 6 h, at a multiplicity of infection (MOI) of 0.1, 72 h post transfer into the hypoxia workstation for Hypo-Diff Huh7.5 cells and 24 h or 72 h post-seeding for Std Huh7.5 cells. The cells were then washed and cultured for an additional 48 h or 72 h in the presence or absence of 1% DMSO, in normoxia or hypoxia, according to the cell culture conditions.
Antibodies
The following antibodies (Ab) were purchased: mouse monoclonal anti-HCV core protein (C7–50, MA1–080, Thermo Fisher Scientific), mouse monoclonal anti-HIF-1α (clone 54, BD Biosciences), rabbit anti-HIF-2α (Ab199, Abcam), rabbit anti-β-actin (ab8227, Abcam, 1:2000), Alexa Fluor™ 594 donkey anti-rabbit IgG (H + L) (Invitrogen), Alexa Fluor™ 594 donkey anti-mouse IgG (H + L) (Invitrogen).
RT-qPCR
Total RNA was extracted with the RNeasy Mini Kit (Qiagen) and viral RNA with the QIAamp Viral RNA Mini Kit (Qiagen), according to the manufacturer’s instructions. Complementary DNA was synthesized with the ProtoScript II First-Strand cDNA Synthesis Kit (New England Biolabs), according to the manufacturer’s instructions. Quantitative PCR was performed with LightCycler 480 SYBR Green Master Mix (Roche Diagnostics). Amplification signals were analyzed with LightCycler 480 software. Gene and viral expression assay information are available in Table 1 (Supplemental Material).
SDS-PAGE and immunoblots
The cells were lysed in RIPA buffer (20 mM Tris, pH 7.5, 20 mM EDTA, 150 mM NaCl, 1% NP40, and 1% sodium deoxycholate) supplemented with protease inhibitor cocktail tablets (Thermo Fisher Scientific). The resulting lysate was centrifuged, and the protein concentration of the supernatant was assessed using the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific). Reducing buffer was added to samples before incubating at 95 °C for 5 min. Forty µg of proteins were separated on 12% polyacrylamide gels and transferred to PVDF membranes (Amersham). Membranes were blocked in PBST, 5% BSA, and proteins detected using specific primary anti-HIF-1α (clone 54; 1:250), anti-HIF-2α (Ab199; 1:500), anti-β-actin (ab8227, Abcam, 1:2000) and corresponding HRP-secondary antibodies. Protein bands were revealed by enhanced chemiluminescence on an Imagequant LAS500 apparatus (GE Healthcare).
Indirect immunofluorescence and lipid droplet staining
Cells were grown on 12-mm glass coverslips and were fixed and permeabilized by incubation with 4% paraformaldehyde in PBS for 10 min at room temperature. After washing three times in PBS, cells were permeabilized and non-specific sites were saturated by incubation for 30 min at room temperature in 0.3% BSA – 0.4% Triton X100 in PBS. Cells were then labeled with an anti-HIF-1α (clone 54; 1:100) or an anti-HIF-2α (Ab199; 1:200) antibody, or a mouse monoclonal anti-HCV core protein (C7-50; 1:250) in PBS-BSA 0.2% by incubation for 1 h at room temperature. The cells were washed three times in PBS and incubated for 1 h at room temperature with an Alexa Fluor™-conjugated secondary antibody (1:2000) in the same buffer. Lipid droplets were stained by incubating cells with Bodipy 493/503 (Fisher Scientific) at a dilution of 1:10,000 in PBS for 5 min at room temperature. Cells were washed three times in PBS and were then mounted in a mixture of equal volumes of Fluoromount-G and DAPI Fluoromount-G (SouthernBiotech). Confocal microscopy was performed with an SP8 confocal microscope (Leica).
The size and number of lipid droplets per cell were analyzed with Image J software (NIH, Bethesda, MD, USA) after being defined and selected, for five representative fields in each set of conditions. Histograms of lipid droplet sizes were generated from data normalized by the number of cells in each image.
Infectivity of the collected supernatants
Fifty µL of fresh supernatant from JFH1-infected Std and HIF-1α or HIF-2α-silenced Hypo-Diff Huh7.5 cells were transferred for 6 h onto 15,000 JFH1-naive Huh7.5 cells, cultured in Std cell culture conditions in 96-well plates. The cells were thoroughly washed with PBS and incubated in complete DMEM for 72 h. Cells were fixed and permeabilized by incubation with cold 50% acetone—50% ethanol (vol/vol) for 10 min at room temperature. Indirect immunofluorescence analyses were then performed, as described above. Focus-forming units (FFU) were counted (Olympus IX 51, Tokyo, Japan), and the results are expressed as FFU/mL/105 HCV genome copies of supernatant from JFH1-infected cells.
Buoyant density on iodixanol gradient ultracentrifugation
Fresh supernatants from JFH1-infected Std and HIF-1α or HIF-2α-silenced Hypo-Diff Huh7.5 cells were overlaid on iodixanol gradients generated from equal volumes (2 mL) of 4, 13, 22, 31 and 40% (weight/weight) Opti-Prep density gradient medium (Merk) solutions in gradient buffer (0.25 M sucrose, 0.5 M Tris pH = 8.0, 0.1 M EDTA pH = 8.8, 2 M MgCl2, 2 M MgSO4). Equilibrium was achieved by ultracentrifugation for 18 h at 250,000 × g in an SW41 rotor at 4 °C in a Beckman Coulter Optima XPN80 ultracentrifuge. Twelve fractions (1 mL each) were collected from the top. The viral genome was quantified in these fractions and the infectivity of the viral particles in each fraction was analyzed as described above. The density of each fraction was determined by measuring its mass.
Statistical analysis
Statistical analyses were performed with Prism software (GraphPad Software Inc., San Diego, California, USA). The results are expressed as the mean ± SEM of at least three sample replicates unless otherwise stated. The statistical significance of differences was assessed in two-tailed Student's t-tests or Welch t-tests, depending on the normality of the data and the homoscedasticity of the variance, as determined by Shapiro–Wilk tests and F tests, respectively. All p values below 0.05 were considered significant.
Results
The silencing approach of HIF-1α and HIF-2α is efficient and specific in hepatocyte-like cells exposed to sustained hypoxia
Our previous work demonstrated the high efficiency to combine both Huh7.5 cells partial differentiation by exposure to DMSO and to culture cells under sustained hypoxia (Hypo-Diff) to enable the production of highly lipidated and infectious HCV-LVPs, compared to standard Huh7.5 cell-culture conditions without DMSO and in normoxia (Std) [12]. Here we have reproduced these experimental conditions to decipher the precise role of HIF-1α and HIF-2α in these viral characteristics using RNA interference (Fig. 1A). Both specificity and efficacy of siRNAs to achieve HIF-1α and HIF-2α down-regulation were controlled by complementary approaches including HIFα mRNA and protein detection (i.e. western blot and immunofluorescence) after adapted transfections four days post hypoxia (Fig. 1A, B and C). Interestingly, HIF-1α and HIF-2α were not detected in Std cell nuclei on immunofluorescence analyses, whereas both transcription factors were found to have an extensive nuclear distribution in Hypo-Diff cells, a sub cellular localization that was lost after specific transfection of siHIF-1α and siHIF-2α (Fig. 1B and C). The functional impact of this interference was confirmed since the quantities of HIF-1-specifically transcribed mRNA of glucose transporter 1 (GLUT1) and phosphoglycerate kinase 1 (PGK1) and of the HIF-2-targeted superoxide dismutase-2 (SOD-2) were dramatically and specifically down-regulated of about 55, 77 and 55% after transfection with siHIF-1α and siHIF-2α in hypoxic conditions, respectively (Fig. 1D and E). The silencing strategy thus shown to be effective to specifically and independently down-regulate HIF-1 and HIF-2 transcription factors.
Fig. 1.
The silencing approach of HIF-1α and HIF-2α and characterization of the efficiency. (A) Schematic diagram of the experimental procedure. Std Huh7.5 cells were de-differentiated control cells cultured for 6 days at high oxygen tension (normoxia - 21% O2); Hypo-Diff Huh7.5 cells were cultured for 9 days with 1% DMSO in normoxia and then for 6 additional days with 1% DMSO in a low oxygen pressure (hypoxia - 1% O2). These cells were transfected or not with HIF-1/2α at day 13 post DMSO-exposure and day 4 of hypoxia. Abbreviations: D-1, day - 1; D0, day 0; D6, day 6; D9, day 9; D15, day 15. (B) Levels of HIF-1α mRNA were measured in triplicate by RT-qPCR in three independent experiments in Std cells, Hypo-Diff cells, siRNA-transfected scramble Hypo-Diff cells, siRNA-transfected HIF-1α Hypo-Diff cells and siRNA-transfected HIF-2α Hypo-Diff cells (left panel). Relative expression levels were obtained by normalization relative to β-actin and compared with those for Std cells. *** P < 0.001, ns = not significant (Welch's t-test). Representative image of immunoblot analysis of endogenous HIF-1α and β-actin proteins (loading control) levels in Std cells and non-transfected and siRNA-transfected Hypo-Diff cells (middle panel). Indirect immunofluorescence for HIF-1α (red) in Std cells and non-transfected and siRNA-transfected Hypo-Diff cells. Nuclei are shown in blue (DAPI). Scale bars = 25 µm (right panel). (C) Levels of HIF-2α mRNA were measured in triplicate by RT-qPCR in three independent experiments in Std cells, Hypo-Diff cells, siRNA-transfected scramble Hypo-Diff cells, siRNA-transfected HIF-1α Hypo-Diff cells and siRNA-transfected HIF-2α Hypo-Diff cells (left panel). Relative expression levels were obtained by normalization relative to β-actin and compared with those for Std cells. *** P < 0.001, ns = not significant (Welch's t-test). Representative image of immunoblot analysis of endogenous HIF-2α and β-actin proteins (loading control) levels in Std cells and non-transfected and siRNA-transfected Hypo-Diff cells (middle panel). Indirect immunofluorescence for HIF-2α (red) in Std cells and non-transfected and siRNA-transfected Hypo-Diff cells. Nuclei are shown in blue (DAPI). Scale bars = 25 µm (right panel). (D) mRNA relative quantities of HIF-1-targeted genes of glucose transporter 1 (GLUT1) and phosphoglycerate kinase 1 (PGK1) measured in triplicate by RT-qPCR in three independent experiments. Relative expression levels were obtained by normalization relative to β-actin and compared with those for Sdt cells. * P < 0.05, ** P < 0.01, *** P < 0.001, (Welch's t-test). (E) mRNA relative quantities of the HIF-2- targeted gene of superoxide dismutase-2 (SOD-2) measured in triplicate by RT-qPCR in three independent experiments. Relative expression levels were obtained by normalization relative to β-actin and compared with those for Std cells. * P < 0.05, *** P < 0.001, (Welch's t-test)
The size and number of cytoplasmic lipid droplets are controlled by HIF-2α in Hypo-Diff Huh7.5 cells
Intracellular neutral lipids are key elements required for HCV LVP assembly, morphogenesis and egress from infected cells [22, 23]. In DMSO-differentiated Huh7.5 cells exposed to a physiological hypoxia, compared to Std cell culture conditions, we previously showed an increase of both the size and number of cytoplasmic lipid droplets (LDs), the organelles responsible for intracellular neutral lipid storage [12]. Using the specific siRNA-mediated knockdown of HIF-1α and HIF-2α we therefore determined their importance in the morphological characteristics of intracellular LDs. Neutral lipids were stained with Bodipy 493/503, images were captured and the number of LDs per cell was determined (Fig. 2A and Fig. 2B). Analyses confirmed that Hypo-Diff Huh7.5 cells contained significantly more LDs than Std-cultured cells (median 14.7 ± 2.7 LDs/cell and 6.7 ± 0.5 LDs/cell, respectively, P < 0.01) (Fig. 2B). Remarkably, while these amounts were unchanged in HIF-1α knockdown Hypo-Diff Huh7.5, the number of LDs per cell significantly decreased 1.7-fold in siRNA HIF-2α-transfected cells compared to control Hypo-Diff Huh7.5. The values obtained for siRNA HIF-2α-transfected cells were comparable to and not statistically different from Std cell culture conditions (median 8.3 ± 1.8 LDs/cell and 6.7 ± 0.5 LDs/cell, respectively, ns). A mean-volume analysis confirmed that LDs contained in Hypo-Diff Huh7.5 cells were significantly larger than in Std cells (3.55 ± 0.48 µm2 and 1.71 ± 0.31 µm2, respectively, P < 0.001) (Fig. 2C). Noteworthy, the down-modulation of HIF-1α in Hypo-Diff Huh7.5 cells further increased, moderately although significantly, these sizes (4.4 ± 0.56 µm2 and 3.55 ± 0.48 µm2, respectively, P < 0.05) further arguing against a role of HIF-1α and suggesting the responsibility of HIF-2α in the morphogenesis of large LDs. HIF-2α-depleted Hypo-Diff cells values largely confirmed this hypothesis since LDs size dramatically and significantly dropped as compared to both HIF-1α siRNA transfected cells and Hypo-Diff counterparts (2.14 ± 0.31 µm2, 4.4 ± 0.56 µm2 and 3.55 ± 0.48 µm2, respectively, P < 0.001) to reach levels similar to Std cell culture conditions (1.71 ± 0.31 µm2).
Fig. 2.
HIF-2α regulates both the number and the size of lipid droplets contained into Hypo-Diff Huh7.5 cells. (A) Representative images of three independent replicates of staining for intracellular lipid droplets and cell nuclei with Bodipy 493/503 (green) and DAPI (blue), respectively in Hypo-Diff cells, siRNA-transfected scramble Hypo-Diff cells, siRNA-transfected HIF-1α Hypo-Diff cells and siRNA-transfected HIF-2α Hypo-Diff cells. Scale bars = 25 µm. Five randomly chosen fields for each condition representing a total of 1980 cells (Std Huh7.5, n = 648; Hypo-Diff Huh7.5, n = 353; scramble siRNA Huh7.5, n = 281; HIF-1α siRNA Huh7.5, n = 307 and HIF-2α siRNA Huh7.5, n = 391) were captured and the number per cell (B) and volume (C) of the 25,943 total lipid droplets (Std Huh7.5, n = 7090; Hypo-Diff Huh7.5, n = 3814; scramble siRNA Huh7.5, n = 4702; HIF-1α siRNA Huh7.5, n = 6005 and HIF-2α siRNA Huh7.5, n = 4332) were determined with Image J software. ** P < 0.01, *** P < 0.001, ns = not significant (Welch’s t-test)
These results indicate that physiological hypoxia and predominantly HIF-2α participate in the generation of neutral lipid storage in the form of LDs in partially differentiated Huh7.5 cells cultured in physiological hypoxia. Given the importance of LDs in the intracellular HCV life cycle, the importance of HIF-2α was therefore explored.
The highly infectious viral particles produced by HCV-infected Hypo-Diff Huh7.5 cells is dependent on HIF-2α
The requirement of LDs to serve as an assembly platform for HCV is well documented [22–24]. We thus explored the susceptibility of HIF-2α (and HIF-1α) down-regulated Hypo-Diff Huh7.5 cells to support HCV infection and the ability of these cells to produce infectious LVPs relative to Std and wild-type Hypo-Diff cell culture conditions. To this end, Std and Hypo-Diff cells were infected with the cell culture-adapted HCV strain JFH1 at a multiplicity of infection (MOI) of 0.1 a day before HIFs siRNA transfection (Fig. 3A). Two days post transfection, neutral lipid-rich LDs together with the HCV core protein were detected by confocal fluorescence microscopy using Bodipy 493/503 and the mouse monoclonal anti-HCV core protein C7-50, respectively (Fig. 3B). The expression of the HCV core protein was comparable between the standard cell culture condition and both untransfected and siHIF-1α-transfected Hypo-Diff cells. However, siHIF-2α-transfected cells showed a significant downregulation, with approximately 25% reduction compared to the other cell culture conditions (Fig. 3B, left of the dotted line and Fig. 3C). Under all cell culture conditions, JFH1-infected cells exhibited an exclusive localization of core proteins at the surface of LDs, that formed ring-like structures with a perinuclear localization (Fig. 3B, right of the dotted line). Particularly, no different core localization was observed in HIF-1α and HIF-2α down-regulated Hypo-Diff Huh7.5 cultures, indicating that the HCV core-LDs association is independent of both transcription factors, associated target genes and the level of neutral lipid storage.
Fig. 3.
The depletion of HIF-2α in JFH1-infected Hypo-Diff cells impairs both HCV replication and the production of infectious viral LVPs. (A) Schematic diagram of the experimental procedure of HCV infection. Hypo-Diff Huh7.5 cells were cultured for 9 days with 1% DMSO in normoxia and then for 6 additional days with 1% DMSO in a low oxygen pressure (hypoxia—1% O2). These cells were infected with the HCV JFH1 strain (MOI 0.1) at day 12 and transfected or not with HIF-1/2α siRNA at day 13 post DMSO exposure and day 4 of hypoxia. Abbreviations: D-1, day − 1; D0, day 0; D9, day 9; D12, day 12; D13, day 13; D15, day 15. (B) Indirect immunofluorescence of the HCV core protein (red) in non-transfected Std cells and siRNA-transfected Hypo-Diff cells. Nuclei are shown in blue (DAPI) and lipid droplets are shown in green (Bodipy 493/503). Merge images at a low magnification are shown to the left of the dotted line (scale bars = 25 µm); Single stainings and merged images at higher magnification are displayed to the right of the dotted line (scale bars = 10 µm). (C) Three randomly chosen fields for each condition from three independent experiments representing a total of 814 cells (Std Huh7.5, n = 197; Hypo-Diff Huh7.5, n = 160; scramble siRNA Huh7.5, n = 160; HIF-1α siRNA Huh7.5, n = 180 and HIF-2α siRNA Huh7.5, n = 117) were captured and the number of the 230 core positive cells (Std Huh7.5, n = 51; Hypo-Diff Huh7.5, n = 57; scramble siRNA Huh7.5, n = 56; HIF-1α siRNA Huh7.5, n = 58 and HIF-2α siRNA Huh7.5, n = 8) were determined with Image J software. *** P < 0.001, ns = not significant (Welch’s t-test). (D) Intracellular and (E) extracellular HCV RNA levels were measured by RT-qPCR, in triplicate, in three independent experiments, two days post HCVcc infection (JFH1, MOI = 0.1). Values are expressed as HCV genome copies/105 cultured cells. ns: not significant, * P < 0.05, ** P < 0.01 (Unpaired two-tailed Student's t-test). (F) Supernatants of HCV-infected cells were collected and used to infect naïve cells. The infectivity of progeny viral particles was determined by indirect immunofluorescence assays on the HCV core protein, with the results expressed as FFU/mL/105 HCV genome copies for tenfold serially dilutions of culture supernatants. ** P < 0.01, ns = not significant (Unpaired two-tailed Student's t-test), FFU = focus-forming units
Two days post-siRNA transfections and three days post-HCV infection, intracellular and extracellular HCV genomes were quantified by RT-qPCR. Due to their long-term exposure to DMSO, Hypo-Diff Huh7.5 cells are almost quiescent, unlike Std cell cultures (data not shown). Thus, to exclude bias in viral load analyses, cells were counted at the time of RNA extraction and the results were standardized per 105 cultured cells. Infections of Hypo-Diff Huh7.5 cells were nearly as efficient as in Std cells since a weak but not significant decrease in the intra- and extracellular HCV genome amounts was measured (Fig. 3D and Fig. 3E) (intracellular: 3.9 × 107 ± 9.7 × 106 HCV RNA copies/105 cells and 8.4 × 107 ± 4.8 × 106 HCV RNA copies/105 cells, respectively, ns; extracellular: 4.4 × 105 ± 6.5 × 104 HCV RNA copies/105 cells and 1.5 × 106 ± 1.9 × 105 HCV RNA copies/105 cells, respectively, ns). These values were close and not statistically different in HIF-1α down-regulated Hypo-Diff and in scramble-siRNA transfected Huh7.5 cells. Intra- and extracellular HCV genome amounts were in contrast dramatically decreased in HIF-2α down-regulated Hypo-Diff cells compared to Std and HIF-1α transfected Huh7.5 cells (intracellular: 5.95 × 105 ± 1.4 × 105 HCV RNA copies/105 cells, 8.4 × 107 ± 4.8 × 106 HCV RNA copies/105 cells, P < 0.001 and 2.8 × 107 ± 9.8 × 106 HCV RNA copies/105 cells, respectively, P < 0.05; extracellular: 2.1 × 104 ± 5 × 103 HCV RNA copies/105 cells, 1.5 × 106 ± 1.9 × 105 HCV RNA copies/105 cells, P < 0.05 and 3.0 × 105 ± 5.3 × 104 HCV RNA copies/105 cells, respectively, P < 0.05). These data, along with those showing lower expression of the core protein in HIF-2α-depleted Hypo-Diff cells (Fig. 3B, left of the dotted line and Fig. 3C), strongly suggest that HIF-2α is a key factor in HCV replication under physiological conditions of oxygenation. HCVcc-containing extracellular medium from JFH1-infected Std and the different Hypo-Diff cultures was incubated with naïve Huh7.5 cells to complete progeny viral infectivity. Since extracellular amounts of HCV genome copies of these different cultures were not equivalent (Fig. 3E), naïve cells were infected with standardized 105 genome copies for each infection, allowing to measure specific infectious viral efficacy. Three days post-infection, the HCV core protein was stained to calculate FFU, and viral infectivity was determined (Fig. 3F). With an equal number of viral genome used for infections performed with the different supernatants, our results show that progeny viruses produced by HCV infected-Hypo-Diff Huh7.5 cells were eightfold more infectious than those produced by JFH1-infected Std Huh7.5 (9456 ± 824 vs. 1159 ± 166 FFU/mL/105 genome copies, respectively, P < 0.01), confirming our previous findings [12]. Transfection of HCV-producing Hypo-Diff cells with scramble or HIF-1α siRNA did not significantly change these major differences. In contrast, progeny viruses produced by HIF-2α down-regulated Hypo-Diff cells lost these high levels of specific infectivity. In that case, values regressed to those generated for viruses produced in Std conditions (1986 ± 1383 vs. 1159 ± 166 FFU/mL/105 genome copies, respectively, ns). These results thus indicate that HIF-2α has a major role for HCV replication, the production and specific infectivity of progeny viruses generated by hepatocyte-like cells exposed to physiological hypoxia.
To confirm the role of HIF-2α independently from other changes associated with hypoxia, oxygen-insensitive HIF-1α and HIF-2α mutated variant proteins (HIF-1α mut and HIF-2α mut, respectively) were used (Fig. 4). These proteins contain a double mutation (P402 A/P564 A in HIF-1α and P405 A/P531 A in HIF-2α) described as responsible for HIF-1α and HIF-2α stabilization in normoxia [19–21]. Huh7.5 cells in normoxia (Std cells) were thus transfected with these constructions and infected a day later with JFH1 at MOI 1, before harvesting both cells and supernatants (Fig. 4A). Immunoblot analyses displayed high levels of HIF-1α mut (Fig. 4B) and HIF-2α mut (Fig. 4C) protein expressions compared to untransfected and pcDNA3-transfected cells and demonstrated both the role of mutated amino acids in the stabilization of HIF-1/2α in Huh7.5 cells and the efficiency of the transfection approach to stably express these mutated transcription factors in normoxia. We therefore assessed their role in the morphological characteristics of intracellular LDs. Neutral lipids were stained with Bodipy 493/503, images were acquired, and the number and size of LDs per cell were quantified (Fig. 4D, E, and F). The analysis clearly showed that while the stabilized expression of HIF-1α in normoxia did not alter either LD characteristic, HIF-2α mut-transfected cells exhibited significantly more and larger LDs compared to the standard cell culture conditions (median 20.5 ± 2.1 LDs/cell vs. 9.7 ± 1.1 LDs/cell, respectively, P < 0.001; 2.18 ± 0.35 µm2 vs. 1.1 ± 0.25 µm2, respectively, P < 0.001). These findings confirm that HIF-2α contributes to the formation of neutral lipid storage in Huh7.5 cells and reveal that its expression is both necessary and sufficient for this process. JFH1-infection of HIF-1α mut and HIF-2α mut expressing Std cells had no effect on intra- and extracellular HCV genome amounts when quantified by RT-qPCR and compared to untransfected cells (Fig. 4G and H). These data indicate that HIF-1α mut and HIF-2α mut do not perturb viral replication and LVPs egress in the condition of (non-physiological) atmospheric oxygen pressure. In contrast, progeny viruses produced by HIF-2α mut-expressing Std cells displayed a significantly twofold higher infectivity compared to the other conditions (15,733 ± 1527 FFU/mL for HIF-2α mut vs. 6933 ± 611 FFU/mL for untransfected Std, 6733 ± 577 FFU/mL for pcDNA3 and 7600 ± 529 FFU/mL for HIF-1α mut, P < 0.001) (Fig. 4I), indicative for qualitative instead of quantitative LVPs modifications under the control of HIF-2α.
Fig. 4.
The stabilization of HIF-2α in JFH1-infected normoxic cells improves the infectivity of progeny HCV LVPs. (A) Schematic diagram of the experimental procedure of HIF-1/2α mut transfections and HCV infection. Standard de-differentiated Huh7.5 cells cultured in normoxia (21% O2) were seeded at 1 X 105 cells/well the day before transfections with HIF-1α mut, HIF-2α mut or pcDNA3 plasmids. Twenty-four hours post transfections, cells were infected with the HCV JFH1 strain (MOI = 1). Cells and supernatant were collected two days after to evaluate HIF-1/2α mut protein expressions, quantities of intracellular and extracellular HCV genome and to complete progeny HCV LVPs infectivity assays on standard cultured Huh7.5 cells. Abbreviations: D-1, day − 1; D0, day 0; D1, day 1; D3, day 3. Representative immunoblot images of HIF-1α (B, upper panel), HIF-2α (C, upper panel) and β-actin protein (B and C, lower panel) expressions in non-transfected standard (Std), pcDNA3-transfected (pc-DNA3), HIF-1α mut-transfected and in HIF-2α mut-transfected Huh7.5 cells in normoxia. (D) Representative images of three independent replicates of staining for intracellular lipid droplets and cell nuclei with Bodipy 493/503 (green) and DAPI (blue), respectively in Std cells, pcDNA3-transfected cells, HIF-1α mut-transfected Std cells and HIF-2α mut-transfected Std cells. Five randomly chosen fields for each condition representing a total of 1067 cells (Std, n = 403; pcDNA3, n = 194; HIF-1α mut-transfected Std cells, n = 240;and HIF-2α mut-transfected Std cells, n = 230) were captured and the number per cell (E) and volume (F) of the 13,666 total lipid droplets (Std, n = 3892; pcDNA3, n = 2302; HIF-1α mut-transfected Std cells, n = 2770 and; HIF-2α mut-transfected Std cells, n = 4702) were determined with Image J software. *** P < 0.001, ns = not significant (Welch’s t-test). Intracellular (G) and extracellular (H) HCV RNA levels were measured by RT-qPCR, in triplicate, in three independent experiments. Values are expressed as HCV genome copies/µg total RNA for intracellular HCV RNA and HCV genome copies/mL for extracellular HCV RNA. ns: not significant (Unpaired two-tailed Student's t-test). (I) Supernatants of HCV-infected cells were collected and used to infect naïve cells. The infectivity of progeny viral particles was determined by indirect immunofluorescence assays on the HCV core protein, with the results expressed as FFU/mL for tenfold serially dilutions of culture supernatants. *** P < 0.001, ns = not significant (Unpaired two-tailed Student's t-test), FFU = focus-forming units
Overall, these results clearly demonstrate that HIF-2α is both necessary and sufficient to produce highly infectious HCV progeny viruses, alongside an increased number and size of intracellular lipid droplets. In our physiological culture model, we demonstrated that HCV infectivity is dependent on morphological characteristics such as low LVPs density [12]. We thus investigated whether the loss of infectivity of viruses produced by HIF-2α-depleted cells was associated with a defect of lipidation in the Hypo-Diff cell culture model.
HIF-2α in Hypo-Diff Huh7.5 cells controls the production of highly lipidated HCV-LVPs and their related broad infectivity
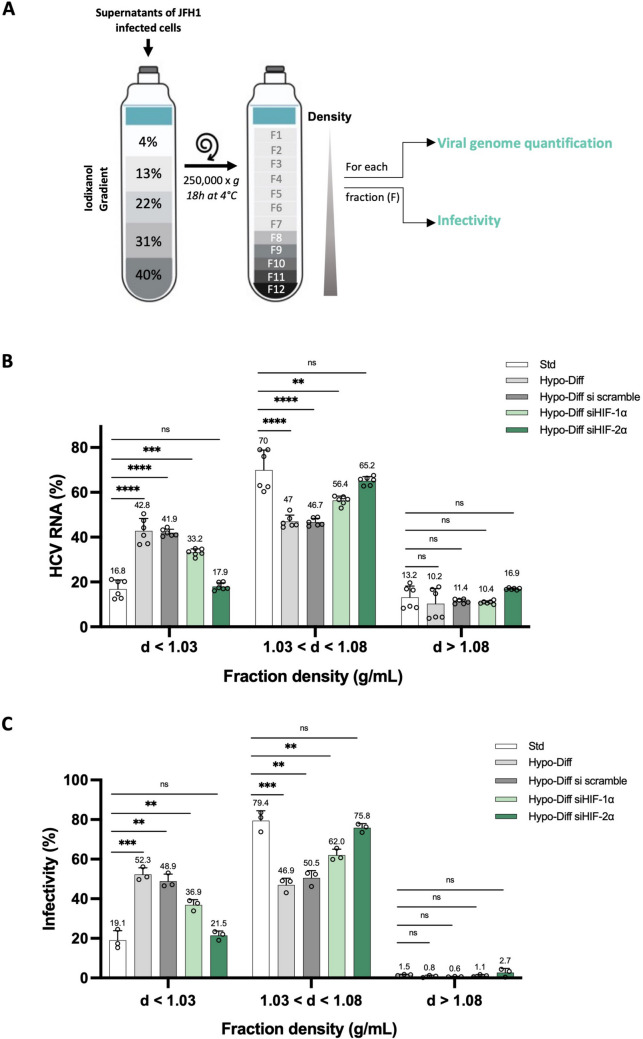
Compared to Std cell culture conditions, HCV-infected Hypo-Diff cells produce highly lipidated and broadly infectious LVPs [12]. As the depletion of HIF-2α in Hypo-Diff cells reduces HCV infectivity, the question then raised as to whether HIF-2α controls the lipidation of HCV LVPs and their associated infectivity. We first determined the relative amounts of HCV RNA present in fractions of various densities isolated from the supernatants of HCV-infected HIF-1α and HIF-2α down-regulated Hypo-Diff cells after iodixanol density gradient centrifugation as summarized in Fig. 5A. As we previously reported [12], we confirmed here that the HCV genome was significantly more abundant for Hypo-Diff cells than for Std cells in low-density fractions (d < 1.03 g/mL) (i.e., highly lipidated LVPs) (42.8 ± 5.6% and 16.8 ± 4%, respectively, P < 0.0001) (Fig. 5B). The depletion of HIF-1α did not significantly change this difference compared to Std cells (P < 0.001). In contrast, the downregulation of HIF-2α in Hypo-Diff cells abrogated the production of highly lipidated HCV LVPs, the density of which was identical to those of Std cell culture conditions (17.9 ± 1.7% and 16.8 ± 4%, respectively, ns). Interestingly, for the density fractions mainly produced by cells (1.03 g/mL < d < 1.08 g/mL), these results were reversed. While Std cells generated a larger proportion of HCV LVPs of lower density (70 ± 8.9%), these proportions dropped for wild-type and HIF-1α down-regulated Hypo-Diff cells (47 ± 2.8% and 56.4 ± 2%, P < 0.0001, P < 0.01, respectively), while they were nearly unchanged for HIF-2α down-regulated Hypo-Diff cells (65.2 ± 1.9%, ns). Thus, overall, HIF-2α-depleted Hypo-Diff cells produce HCV LVPs with a density close to Std cells and significantly higher (i.e., lower lipidated LVPs) than those produced by wild-type and HIF-1α down-regulated Hypo-Diff cells.
Fig. 5.
HIF-2α is responsible for the production of highly lipidated and infectious HCV LVPs by Hypo-Diff Huh7.5 cells. (A) Schematic representation of the experimental procedure for cell culture fractionation and subsequent determination of viral genome quantification and infectivity. Fresh supernatants from JFH1-infected Std and HIF-1α-silenced or HIF-2α-silenced Hypo-Diff Huh7.5 cells were fractioned on iodixanol gradients. Twelve fractions (F1 to F12) were weighted to determine each density together with viral RNA levels and infectivity. (B) HCV RNA was quantified by RT-qPCR on each fraction harvested and the results are expressed as a % of total HCV RNA. ** P < 0.01, *** P < 0.001, **** P < 0.0001, ns: not significant. (Unpaired two-tailed Student's t-test). (C) The infectivity of each of collected fractions was determined with naïve cells and indirect immunofluorescence assays for the HCV core protein. FFU were counted and the results are expressed as the % infectivity (FFU/mL) of fractions with densities < 1.03 g/mL, between 1.03 and 1.08 g/mL and > 1.08 g/mL. ** P < 0.01, *** P < 0.001, ns: not significant (Unpaired two-tailed Student's t-test)
We then determined the infectivity of these fractions, using naïve Huh7.5 cells to calculate FFU/mL and % of viral infectivity (Fig. 5C). Our results confirmed a clear relationship between HCV RNA-associated low buoyant density fractions and the associated infectivity. The proportion of infectious progeny viruses produced by Hypo-Diff, siRNA scramble-transfected and HIF-1α-depleted Hypo-Diff cells associated with the lowest buoyant density fractions (d < 1.03 g/mL) were significantly greater than those produced by Std cells (52.3 ± 3.4%, 48.9 ± 3.5%, 36.9% ± 2.6 and 19.1 ± 4.8%, P < 0.001 and P < 0.01, respectively). These proportions were inverted within higher densities (1.03 < d < 1.08 g/mL) (i.e., lower lipidated LVPs), confirming that Hypo-Diff cells produce significantly more lipidated and infectious HCV LVPs than Std conditions and suggesting that HIF-1α is not a major actor of this feature. In contrast, HCV LVPs produced by HIF-2α-depleted Hypo-Diff cells presented an infectivity identical to that of Std conditions both in lowest buoyant density fractions (d < 1.03 g/mL) (21.5 ± 2.2% vs 19.1 ± 4.8%, ns, respectively) and in higher densities (1.03 < d < 1.08 g/mL) (75.8 ± 2.1% vs 79.4 ± 5%, ns, respectively).
Overall, these results provide a demonstration that HIF-2α controls both the production and infectivity of very low mean buoyant density (i.e. highly lipidated) HCV LVPs from cells that combine two key physiological characteristics observed during natural intrahepatic HCV infection (i.e., cellular differentiation and prolonged hypoxia).
Discussion
HCV viral assembly and morphogenesis, which has been reported to be closely linked to cellular lipid metabolism, remain poorly characterized. In standard cell culture conditions (i.e. de-differentiated cells in normoxia) HCVcc virions display a number of major biochemical and ultrastructural differences with respect to the viral particles that circulate in the blood of HCV-infected patients [4, 6, 7]. Native HCV is actually unique in its ability to associate closely with lipoprotein components, forming hybrid particles known as lipo-viro-particles (LVPs) [9–11]. Our previous study demonstrated that a partial differentiation of HCV-infected Huh7.5 cells cultured in a physiological prolonged hypoxia produces highly infectious progeny viruses displaying strong biochemical and ultrastructural similarities to native LVPs [12]. We confirm here these data but also demonstrate that hypoxia and notably HIF-2α plays a pivotal role to yield these characteristics, in close relation with the storage of neutral lipids within the LVPs-producing cells.
The respective roles of HIF-1α and HIF-2α in the hypoxia-induced hepatic liver metabolism is still debated. HIF-1α would upregulate the expression of genes related to lipid uptake and synthesis in the liver and downregulate the expression of lipid oxidation genes. Thus, it would promote intrahepatic lipid deposition ([25] for review). The role of HIF-2α is also well documented although the explanation of the inductive mechanisms is still controversial. Indeed, while some investigators argue both with a mouse model and HepG2 cell cultures that hepatic HIF-2α overexpression increased fatty acid synthesis and caused severe steatohepatitis [26, 27], others defend that HIF-2α-induced lipid accumulation in the liver is due to impaired fatty acid β-oxidation rather than upregulation of lipogenic genes [16]. We demonstrate here that HIF-2α, but not HIF-1α, is responsible for the accumulation of large neutral lipid-rich droplets within partially differentiated Huh7.5 cells cultured in a physiological prolonged hypoxia (Fig. 2). Whether this storage is caused by an increased lipid synthesis or a default in fatty acid β-oxidation is an important issue that will need to be investigated. Lipid droplets are known to serve as an assembly platform for HCV, notably characterized by a close association with the HCV core protein [22, 23]. In JFH1 infected cultures, HIF-2α-depleted cells did not exhibit a disruption of this characteristic (Fig. 3B) suggesting a disconnection between HIF-2α-dependent neutral lipid storage and HCV core ability to appropriately associate with LDs for viral assembly. Among other LDs-associated proteins, diacylglycerol acyltransferase 1 (DGAT 1), a rate-limiting enzyme in triglyceride synthesis from diacylglycerol, is required for the binding of HCV core protein but also for NS3 to LDs [28]. In HIF-2α-depleted cells exhibiting a default of neutral lipid storage, DGAT 1 could be supposed to be normally expressed and efficient for HCV-LDs association suggesting that the DGAT 1 promoter would not be targeted by HIFs and particularly by HIF-2.
The combination of physiological hypoxia and Huh7.5 cells differentiation presents neither an advantage nor a barrier to the smooth running of the viral cycle, according to the quantifications of the intra- and extracellular viral genome obtained here and confirming data of our previous work (Fig. 3C and D) [12]. In contrast, HIF-2α-depleted cells exhibited an overall significant viral cycle disruption, highlighting its importance in hypoxia-dependent replication of HCV. Replication is a complex process of the viral life cycle that is facilitated by interaction of HCV with various lipid-related factors ([29] for review). As an example, increasing evidence present unesterified/free cholesterol and sphingolipids as major actors of HCV replication [30, 31]. However, although standard cells and HIF-2α-depleted Hypo-Diff cells exhibit similar characteristics in terms of lipid droplet number and size (Fig. 2), viral replication is significantly reduced in the latter (Fig. 3). Therefore, the regulation of HCV replication by HIF-2α appears to be independent of intracellular storage in neutral lipids, and its mechanism should be investigated through alternative pathways. In addition, HIF-2α-dependent genome replication and viral morphogenesis have to be considered separately. Interestingly our results obtained with the ectopic expression of the HIF-2α mutant stabilized in JFH1-infected standard cells clearly distinguished stable viral genome replication, despite the significant increase of lipid droplet number and size, and the high level of infectivity of progeny LVPs (Fig. 4). These data thus indicate that HIF-2α plays a direct role in the production of broadly infectious LVPs, independently of viral replication.
Our study indeed strongly suggests that the global weakness of the lipid metabolism in HIF-2α-depleted Hypo-Diff cells is individually responsible for the lack of native-like progeny LVPs morphogenesis and their associated weak infectivity (Figs. 3, 4 and 5). The precise mechanism controlled by HIF-2α to modulate the cellular lipid homeostasis is still unknown. While the transcription factor function of HIF-2α is now well established, HIF-2α also operates as a cap-dependent translation initiation factor in oxygen deprived cells [32]. Hypoxia indeed promotes the formation of a translation initiation complex that includes HIF-2α, RNA binding protein RBM4 and eukaryotic initiation factor 4E2 (eIF4E2), only active in hypoxia [33]. Therefore, deciphering the role of HIF-2α in the morphogenesis of HCV LVPs should be approached with great caution regarding the complex cellular regulations controlled by this central actor in hypoxia.
In conclusion, we demonstrate here that physiological hypoxia and, notably, HIF-2α are responsible for intracellular neutral lipid storage and the production of highly lipidated and broadly infectious LVPs in our recently developed HCV-infected cell culture model. These data undoubtedly provide new insights into the understanding of HCV morphogenesis and its associated infectivity during natural intrahepatic HCV infection.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Takaji Wakita (National Institute of Infectious Disease, Tokyo, Japan) for kindly providing the JFH1 virus strain, Charles M. Rice (Rockefeller University, New York, USA) for the Huh7.5 cell line and William C Kaelin (Harvard medical school, Boston, USA) for plasmids expressing stabilized HIF-1α and HIF-2α. We are grateful to Lorena Tomé-Poderti and Florian Seigneuret for fruitful discussions.
Abbreviations
- DAA
Direct acting antiviral
- DGAT 1
Diacylglycerol acyltransferase 1
- FFU
Focus forming units
- GLUT1
Glucose transporter 1
- HCVcc
Hepatitis C virus in cell culture
- HIF
Hypoxia-inducible factor
- JFH1
Japanese fulminant hepatitis
- LD
Lipid droplet
- LVP
Lipo-viro-particle
- PGK1
Phosphoglycerate kinase
- PHD
Prolyl hydroxylase domain
- SOD-2
Superoxyde dismutase-2
- VHL
Von Hippel-Lindau
Author contributions
Design and execution of the experiments, data acquisition, analysis and interpretation were performed by Mathilde Couteaudier. Technical assistance was performed by Mona Nivard and Jade Cochard. A critical revision of the paper was performed by Fabrizio Mammano, Philippe Roingeard and Hugues de Rocquigny. The study design, data analysis and interpretation were performed by Philippe Chouteau. The manuscript was written by Philippe Chouteau. Fundings were obtained by Philippe Chouteau. All authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by a grant from the Agence Nationale de Recherche sur le SIDA et les virus des hépatites/Maladies infectieuses émergentes (ANRS-MIE – ECTZ118678). M.C. was recipient of a grant from ANRS-MIE (ECTZ205976).
Data availability
All data generated or analyzed in the current study are available from the corresponding author on reasonable request.
Declarations
Ethical approval
Not applicable.
Consent for publication
Not applicable.
Consent to participate
Not applicable.
Competing interest
The authors have no relevant financial or non-financial interests to disclose.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Vo-Quang E, Pawlotsky J-M (2024) ‘Unusual’ HCV genotype subtypes: origin, distribution, sensitivity to direct-acting antiviral drugs and behaviour on antiviral treatment and retreatment. 10.1136/gutjnl-2024-332177 [DOI] [PMC free article] [PubMed]
- 2.Childs K, Davis C, Cannon M et al (2019) Suboptimal SVR rates in African patients with atypical genotype 1 subtypes: implications for global elimination of hepatitis C. J Hepatol 71:1099–1105. 10.1016/j.jhep.2019.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fourati S, Rodriguez C, Hézode C et al (2019) Frequent antiviral treatment failures in patients infected with hepatitis C virus genotype 4, subtype 4r. Hepatology 69:513–523. 10.1002/hep.30225 [DOI] [PubMed] [Google Scholar]
- 4.Lindenbach BD, Evans MJ, Syder AJ et al (2005) Complete replication of hepatitis C virus in cell culture. Science 309:623–626. 10.1126/science.1114016 [DOI] [PubMed] [Google Scholar]
- 5.Wakita T, Pietschmann T, Kato T et al (2005) Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med 11:791–796. 10.1038/nm1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Podevin P, Carpentier A, Pène V et al (2010) Production of infectious hepatitis C virus in primary cultures of human adult hepatocytes. Gastroenterology 139:1355–1364. 10.1053/j.gastro.2010.06.058 [DOI] [PubMed] [Google Scholar]
- 7.Nielsen SU, Bassendine MF, Burt AD et al (2006) Association between hepatitis C virus and very-low-density lipoprotein (VLDL)/LDL analyzed in iodixanol density gradients. J Virol 80:2418–2428. 10.1128/JVI.80.5.2418-2428.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Piver E, Boyer A, Gaillard J et al (2017) Ultrastructural organisation of HCV from the bloodstream of infected patients revealed by electron microscopy after specific immunocapture. Gut 66:1487–1495. 10.1136/gutjnl-2016-311726 [DOI] [PubMed] [Google Scholar]
- 9.Vieyres G, Pietschmann T (2019) HCV pit stop at the lipid droplet: refuel lipids and put on a lipoprotein coat before exit. Cells 8:233. 10.3390/cells8030233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.André P, Komurian-Pradel F, Deforges S et al (2002) Characterization of low- and very-low-density hepatitis C virus RNA-containing particles. J Virol 76:6919–6928. 10.1128/jvi.76.14.6919-6928.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Catanese MT, Uryu K, Kopp M et al (2013) Ultrastructural analysis of hepatitis C virus particles. Proc Natl Acad Sci U S A 110:9505–9510. 10.1073/pnas.1307527110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cochard J, Bull-Maurer A, Tauber C et al (2021) Differentiated cells in prolonged hypoxia produce highly infectious native-like hepatitis C virus particles. Hepatology 74:627–640. 10.1002/hep.31788 [DOI] [PubMed] [Google Scholar]
- 13.Solanki S, Shah YM (2024) Hypoxia-induced signaling in gut and liver pathobiology. Annu Rev Pathol 19:291–317. 10.1146/annurev-pathmechdis-051122-094743 [DOI] [PubMed] [Google Scholar]
- 14.Strowitzki MJ, Cummins EP, Taylor CT (2019) Protein hydroxylation by hypoxia-inducible factor (HIF) hydroxylases: unique or ubiquitous? Cells 8:384. 10.3390/cells8050384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taniguchi CM, Finger EC, Krieg AJ et al (2013) Cross-talk between hypoxia and insulin signaling through Phd3 regulates hepatic glucose and lipid metabolism and ameliorates diabetes. Nat Med 19:1325–1330. 10.1038/nm.3294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rankin EB, Rha J, Selak MA et al (2009) Hypoxia-inducible factor 2 regulates hepatic lipid metabolism. Mol Cell Biol 29:4527–4538. 10.1128/MCB.00200-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goda N, Kanai M (2012) Hypoxia-inducible factors and their roles in energy metabolism. Int J Hematol 95:457–463. 10.1007/s12185-012-1069-y [DOI] [PubMed] [Google Scholar]
- 18.Yan Q, Bartz S, Mao M et al (2007) The hypoxia-inducible factor 2alpha N-terminal and C-terminal transactivation domains cooperate to promote renal tumorigenesis in vivo. Mol Cell Biol 27:2092–2102. 10.1128/MCB.01514-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kondo K, Kim WY, Lechpammer M, Kaelin WG (2003) Inhibition of HIF2α is sufficient to suppress pVHL-defective tumor growth. PLoS Biol. 10.1371/journal.pbio.0000083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee K, Kang JE, Park S-K et al (2010) LW6, a novel HIF-1 inhibitor, promotes proteasomal degradation of HIF-1α via upregulation of VHL in a colon cancer cell line. Biochem Pharmacol 80:982–989. 10.1016/j.bcp.2010.06.018 [DOI] [PubMed] [Google Scholar]
- 21.Hashimoto T, Shibasaki F (2015) Hypoxia-inducible factor as an angiogenic master switch. Front Pediatr 3:33. 10.3389/fped.2015.00033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miyanari Y, Atsuzawa K, Usuda N et al (2007) The lipid droplet is an important organelle for hepatitis C virus production. Nat Cell Biol 9:1089–1097. 10.1038/ncb1631 [DOI] [PubMed] [Google Scholar]
- 23.Boulant S, Targett-Adams P, McLauchlan J (2007) Disrupting the association of hepatitis C virus core protein with lipid droplets correlates with a loss in production of infectious virus. J Gen Virol 88:2204–2213. 10.1099/vir.0.82898-0 [DOI] [PubMed] [Google Scholar]
- 24.Galli A, Scheel TKH, Prentoe JC et al (2013) Analysis of hepatitis C virus core/NS5A protein co-localization using novel cell culture systems expressing core-NS2 and NS5A of genotypes 1–7. J Gen Virol 94:2221–2235. 10.1099/vir.0.053868-0 [DOI] [PubMed] [Google Scholar]
- 25.Luo M, Li T, Sang H (2023) The role of hypoxia-inducible factor 1α in hepatic lipid metabolism. J Mol Med (Berl) 101:487–500. 10.1007/s00109-023-02308-5 [DOI] [PubMed] [Google Scholar]
- 26.Rey E, Meléndez-Rodríguez F, Marañón P et al (2020) Hypoxia-inducible factor 2α drives hepatosteatosis through the fatty acid translocase CD36. Liver Int 40:2553–2567. 10.1111/liv.14519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qu A, Taylor M, Xue X et al (2011) Hypoxia-inducible transcription factor 2α promotes steatohepatitis through augmenting lipid accumulation, inflammation and fibrosis. Hepatology 54:472–483. 10.1002/hep.24400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herker E, Harris C, Hernandez C et al (2010) Efficient hepatitis C virus particle formation requires diacylglycerol acyltransferase 1 (DGAT1). Nat Med 16:1295–1298. 10.1038/nm.2238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sidorkiewicz M (2021) Hepatitis C virus uses host lipids to its own advantage. Metabolites 11:273. 10.3390/metabo11050273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aizaki H, Lee K-J, Sung VM-H et al (2004) Characterization of the hepatitis C virus RNA replication complex associated with lipid rafts. Virology 324:450–461. 10.1016/j.virol.2004.03.034 [DOI] [PubMed] [Google Scholar]
- 31.Shi ST, Lee K-J, Aizaki H et al (2003) Hepatitis C virus RNA replication occurs on a detergent-resistant membrane that cofractionates with caveolin-2. J Virol 77:4160–4168. 10.1128/JVI.77.7.4160-4168.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Uniacke J, Holterman CE, Lachance G et al (2012) An oxygen-regulated switch in the protein synthesis machinery. Nature 486:126–129. 10.1038/nature11055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uniacke J, Kishan Perera J, Lachance G et al (2014) Cancer cells exploit eIF4E2-directed synthesis of hypoxia response proteins to drive tumor progression. Can Res 74:1379–1389. 10.1158/0008-5472.CAN-13-2278 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed in the current study are available from the corresponding author on reasonable request.