Extending lifespan with Hadoop and R
- 1. Extending lifespan with R and Hadoop Radek Maciaszek Founder of DataMine Lab, CTO Ad4Game, studying towards PhD in Bioinformatics at UCL
- 2. Agenda ● Project background ● Parallel computing in R ● Hadoop + R ● Future work (Storm) ● Results and summary 2
- 3. Project background ● Lifespan extension - project at UCL during MSc in Bioinformatics ● Bioinformatics – computer science in biology (DNA, Proteins, Drug discovery, etc.) ● Institute of Healthy Ageing at UCL – lifespan is a king. Dozens of scientists, dedicated journals. ● Ageing is a complex process or is it? C. Elegans (2x by a single gene DAF-2, 10x). ● Goal of the project: find genes responsible for ageing 3 Caenorhabditis Elegans
- 4. Primer in Bioinformatics ● Central dogma of molecular biology ● Cell (OS+3D), Gene (Program), TF (head on HDD) ● How to find ageing genes (such as DAF-2)? 4 Images: Wikipedia
- 5. RNA microarray DAF-2 pathway in C. elegans Source: Partridge & Gems, 2002 Source: Staal et al, 2003 5
- 6. Goal: raw data → network Genes Network ● Pairwise comparisons of 10k x 10k genes + clustering 100 x 100 x 50 x 10 (~10k genes) 6
- 7. Why R? ● Incredibly powerful for data science with big data ● Functional, scripting programming language with many packages. ● Popular in mathematics, bioinformatics, finance, social science and more. ● TechCrunch lists R as trendy technology for BigData. ● Designed by statisticians for statisticians 7
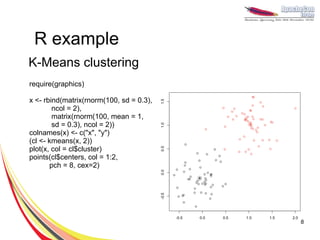
- 8. R example K-Means clustering require(graphics) x <- rbind(matrix(rnorm(100, sd = 0.3), ncol = 2), matrix(rnorm(100, mean = 1, sd = 0.3), ncol = 2)) colnames(x) <- c("x", "y") (cl <- kmeans(x, 2)) plot(x, col = cl$cluster) points(cl$centers, col = 1:2, pch = 8, cex=2) 8
- 9. R limitations & Hadoop ● 10k x 10k (100MM) Fisher exact correlations is slow ● Memory allocation is a common problem ● Single-threaded ● Hadoop integration: – Hadoop Streaming – Rhipe: http://ml.stat.purdue.edu/rhipe/ – Segue: http://code.google.com/p/segue/ 9
- 10. Scaling R ● Explicit – snow, parallel, foreach ● Implicit – multicore (2.14.0) ● Hadoop – RHIPE, rmr, Segue, RHadoop ● Storage – rhbase, rredis, Rcassandra, rhdfs 10
- 11. R and Hadoop ● Streaming API (low level) mapper.R #!/usr/bin/env Rscript in <- file(“stdin”, “r”) while (TRUE) { lineStr <- readLines(in, n=1) line <- unlist(strsplit(line, “,”)) ret = expensiveCalculations(line) cat(data, “n”, sep=””) } close(in) jar hadoop-streaming-*.jar –input data.csv –output data.out –mapper mapper.R 11
- 12. RHIPE ● Can use with your Hadoop cluster ● Write mappers/reduces using R only map <- expression({ z <- f <- table(unlist(strsplit(unlist( rhmr(map=map,reduce=reduce, map.values)," "))) inout=c("text","sequence") n <- names(f) ,ifolder=filename p <- as.numeric(f) ,ofolder=sprintf("%s- sapply(seq_along(n),function(r) out",filename)) rhcollect(n[r],p[r])) }) job.result <- rhstatus(rhex(z,async=TRUE), reduce <- expression( mon.sec=2) pre={ total <- 0}, reduce = { total <- total+sum(unlist(reduce.values)) }, post = { rhcollect(reduce.key,total) } ) 12 Example from Rhipe Wiki
- 13. Segue ● Works with Amazon Elastic MapReduce. ● Creates a cluster for you. ● Designed for Big Computations (rather than Big Data) ● Implements a cloud version of lapply() ● Parallelization in 2 lines of code! ● Allowed us to speed up calculations down to 2h with the use of 16 servers 13
- 14. Segue workflow (emrlapply) 14
- 15. lapply() m <- list(a = 1:10, b = exp(-3:3)) lapply(m, mean)$a [1] 5.5 $b [1] 4.535125 lapply(X, FUN) returns a list of the same length as X, each element of which is the result of applying FUN to the corresponding element of X. 15
- 16. Segue in a cluster > AnalysePearsonCorelation <- function(probe) { A.vector <- experiments.matrix[probe,] p.values <- c() for(probe.name in rownames(experiments.matrix)) { B.vector <- experiments.matrix[probe.name,] p.values <- c(p.values, cor.test(A.vector, B.vector)$p.value) } return (p.values) } > # pearson.cor <- lapply(probes, AnalysePearsonCorelation) Moving to the cloud in 3 lines of code! 16
- 17. Segue in a cluster > AnalysePearsonCorelation <- function(probe) { A.vector <- experiments.matrix[probe,] p.values <- c() for(probe.name in rownames(experiments.matrix)) { B.vector <- experiments.matrix[probe.name,] p.values <- c(p.values, cor.test(A.vector, B.vector)$p.value) } return (p.values) } > # pearson.cor <- lapply(probes, AnalysePearsonCorelation) > myCluster <- createCluster(numInstances=5, masterBidPrice="0.68”, slaveBidPrice="0.68”, masterInstanceType=”c1.xlarge”, slaveInstanceType=”c1.xlarge”, copy.image=TRUE) > pearson.cor <- emrlapply(myCluster, probes, AnalysePearsonCorelation) > stopCluster(myCluster) 17
- 18. R + HBase library(rhbase) hb.init(serialize="raw") #create new table hb.new.table("mytable", "x","y","z",opts=list(y=list(compression='GZ'))) #insert some values into the table hb.insert("mytable",list( list(1,c("x","y","z"),list("apple","berry","cherry")))) rows<-hb.scan.ex("mytable",filterstring="ValueFilter(=,'substring:ber')") rows$get() https://github.com/RevolutionAnalytics/RHadoop/wiki/rhbase 18
- 19. Discovering genes Topomaps of clustered genes This work was based on:A Gene Expression Map for Caenorhabditis elegans, Stuart K. Kim, et al., Science 293, 2087 (2001) 19
- 20. Genes clusters Clusters based on Fisher exactpairwise genes comparisons Green lines represent random probes Red lines represent up-regulated probes Blue lines are down-regulated probes (in daf-2 vs daf-2;daf-16 experiment) 20
- 21. Genes networks Network created with Cytoscape, platform for complex network analysis: http://www.cytoscape.org/ 21
- 22. Future work - real time R ● Hadoop has high throughput but for small tasks is slow. It is not good for continuous calculations. ● A possible solution is to use Storm ● Storm multilang can be used with any language, including R 22
- 23. Storm R Storm may be easily integrated with third party languages and databases: ● Java ● Python ● Ruby ● Redis ● Hbase ● Cassandra Image source: Storm github wiki 23
- 24. Storm R source("storm.R") initialize <- function() { emitBolt(list("bolt initializing")) } process <- function(tup) { word <- tup$tuple rand <- runif(1) if (rand < 0.75) { emitBolt(list(word + "lalala")) } else { log(word + " randomly skipped!") } } boltRun(process, initialize) https://github.com/rathko/storm 24
- 25. Summary ● It’s easy to scale R using Hadoop. ● R is not only great for statistics, it is a versatile programming language. ● Is ageing a disease? Are we all going to live very long lives? 25
- 26. Questions? ● References: http://hadoop.apache.org/ http://hbase.apache.org/ http://code.google.com/p/segue/ http://www.datadr.org/ https://github.com/RevolutionAnalytics/ https://github.com/rathko/storm 26











![RHIPE
● Can use with your Hadoop cluster
● Write mappers/reduces using R only
map <- expression({
z <- f <- table(unlist(strsplit(unlist(
rhmr(map=map,reduce=reduce, map.values)," ")))
inout=c("text","sequence") n <- names(f)
,ifolder=filename p <- as.numeric(f)
,ofolder=sprintf("%s- sapply(seq_along(n),function(r)
out",filename)) rhcollect(n[r],p[r]))
})
job.result <-
rhstatus(rhex(z,async=TRUE), reduce <- expression(
mon.sec=2) pre={ total <- 0},
reduce = { total <-
total+sum(unlist(reduce.values)) },
post = { rhcollect(reduce.key,total) }
)
12
Example from Rhipe Wiki](https://image.slidesharecdn.com/rhadoopoo-121107053033-phpapp01/85/Extending-lifespan-with-Hadoop-and-R-12-320.jpg)


![lapply()
m <- list(a = 1:10, b = exp(-3:3))
lapply(m, mean)$a
[1] 5.5
$b
[1] 4.535125
lapply(X, FUN)
returns a list of the same length as X, each element of which is
the result of applying FUN to the corresponding element of X.
15](https://image.slidesharecdn.com/rhadoopoo-121107053033-phpapp01/85/Extending-lifespan-with-Hadoop-and-R-15-320.jpg)
![Segue in a cluster
> AnalysePearsonCorelation <- function(probe) {
A.vector <- experiments.matrix[probe,]
p.values <- c()
for(probe.name in rownames(experiments.matrix)) {
B.vector <- experiments.matrix[probe.name,]
p.values <- c(p.values, cor.test(A.vector, B.vector)$p.value)
}
return (p.values)
}
> # pearson.cor <- lapply(probes, AnalysePearsonCorelation)
Moving to the cloud in 3 lines of code!
16](https://image.slidesharecdn.com/rhadoopoo-121107053033-phpapp01/85/Extending-lifespan-with-Hadoop-and-R-16-320.jpg)
![Segue in a cluster
> AnalysePearsonCorelation <- function(probe) {
A.vector <- experiments.matrix[probe,]
p.values <- c()
for(probe.name in rownames(experiments.matrix)) {
B.vector <- experiments.matrix[probe.name,]
p.values <- c(p.values, cor.test(A.vector, B.vector)$p.value)
}
return (p.values)
}
> # pearson.cor <- lapply(probes, AnalysePearsonCorelation)
> myCluster <- createCluster(numInstances=5,
masterBidPrice="0.68”, slaveBidPrice="0.68”,
masterInstanceType=”c1.xlarge”,
slaveInstanceType=”c1.xlarge”, copy.image=TRUE)
> pearson.cor <- emrlapply(myCluster, probes,
AnalysePearsonCorelation)
> stopCluster(myCluster)
17](https://image.slidesharecdn.com/rhadoopoo-121107053033-phpapp01/85/Extending-lifespan-with-Hadoop-and-R-17-320.jpg)








