Bio ontologies and semantic technologies[2]
- 1. Introduction to Bio Ontologies and The Semantic Web M. Devisscher Biological Databases
- 2. Overview • Bio ontologies • Semantic technologies • SPARQL in practice
- 3. Introduction • Ontologies: what are ontologies ? • Ontologies in the bio domain: OBO Foundry • Ontologies in the semantic web • OBO • RDF, IRI, TTL, SPARQL, OWL
- 4. What is an ontology ? • Ontology = a specification of a conceptualization (Gruber 1993) • In practice: controlled vocabularies – Disambiguation (e.g. Bank, Running) – Language/species independence • Very useful in biology – complex hierarchies of terms
- 5. Ontologies in the bio Domain • OBO Foundry - open Biological and Biomedical Ontologies • Common principles • List of ontologies at http://www.obofoundry.org • OBO is also a data format .obo
- 6. SideTrack – The Gene Ontology • The mother of bio-ontologies: the GO – Oldest bio – ontology – Many practical applications: • Cross species studies • Overrepresentation studies (RNASeq) • GO is an OBO ontology
- 7. SideTrack – The Gene Ontology • Collection of terms
- 8. SideTrack – The Gene Ontology • Relationships between terms: – Subsumption: is_a – Partonomic: part_of • These terms are transitive • Terms form a DAG (directed, acyclic graph) • Some information can be inferred
- 11. SideTrack – The Gene Ontology • Know more: www.geneontology.org • AMIGO : the GO browser
- 13. Semantic Technologies • The semantic web: Tim Berners Lee et al, Scientific American 2001
- 14. Semantic Technologies • W3C: a set of specifications http://www.w3.org/standards/semanticweb/ • A mature toolset – Dedicated data formats – Storage – Query language
- 15. Resource Description Framework • A standard model for data interchange on the (semantic) web • Basic data element = a Triple – A mini sentence – Contains three Terms: • Subject Predicate Object
- 16. • Representation of triples – Basic data format: RDF/XML – All data expressed in RDF (Resource Description Framework) – Several compatible syntaxes: TTL (Terse Triple Language) most human readable Resource Description Framework
- 17. Example
- 19. The Turtle Syntax • Prefix @prefix b4x: <http:bioinformatics.be/terms#> b4x:martijn b4x:has_favorite_beer b4x:karmeliet.
- 20. The Turtle Syntax • Predicate lists @prefix b4x: <http:bioinformatics.be/terms#> . @prefix foaf: <http://xmlns.com/foaf/0.1/> . b4x:martijn b4x:has_favorite_beer b4x:karmeliet; foaf:name “Martijn Devisscher”.
- 21. The Turtle Syntax • Object lists @prefix b4x: <http:bioinformatics.be/terms#> . @prefix foaf: <http://xmlns.com/foaf/0.1/> . b4x:martijn b4x:has_favorite_beer b4x:karmeliet, b4x:chimay_blauw; foaf:name “Martijn Devisscher”.
- 22. IRI’s and Literals • Terms can be either IRI’s, Literals or blank nodes • IRI = Internationalized Resource Identifier • Unique id – a virtual URI – Example: <http://bioinformatics.be/terms#martijn> – There is no requirement for resolving – Now: Open Data initiatives: please do use resolvable URI’s http://linkeddata.org – Unique identifiers can be registered on http://identifiers.org
- 23. Introduction • Literals: can be typed, allowed types from the XSD namespace: – E.g. “This is a string example”^^xsd:string – E.g. “5”^^xsd:integer • IRI’s are used for entities and attributes • Literals are used for attribute values that aren’t entities
- 24. The Turtle Syntax • Typed literals @prefix b4x: <http:bioinformatics.be/terms#> . @prefix foaf: <http://xmlns.com/foaf/0.1/> . @prefix xsd: <http://www.w3.org/2001/XMLSchema#> . b4x:martijn b4x:has_favorite_beer b4x:karmeliet, b4x:chimay_blauw; b4x:length “184”^^xsd:integer; foaf:name “Martijn Devisscher”^^xsd:string.
- 25. The Turtle Syntax • Blank nodes @prefix b4x: <http:bioinformatics.be/terms#> . @prefix foaf: <http://xmlns.com/foaf/0.1/> . @prefix xsd: <http://www.w3.org/2001/XMLSchema#> . b4x:martijn b4x:has_favorite_beer b4x:karmeliet, b4x:chimay_blauw; b4x:length “184”^^xsd:integer; foaf:name “Martijn Devisscher”^^xsd:string; b4x:owns_cat [ b4x:color “Gray” ].
- 26. Classes and Individuals • rdf:type @prefix b4x: <http:bioinformatics.be/terms#> . @prefix foaf: <http://xmlns.com/foaf/0.1/> . b4x:martijn rdf:type foaf:Person.
- 27. Classes and Individuals • Shorthand: a @prefix b4x: <http:bioinformatics.be/terms#> . @prefix foaf: <http://xmlns.com/foaf/0.1/> . b4x:martijn a foaf:Person; foaf:knows b4x:geert. b4x:geert a foaf:Person.
- 29. Semantic Technologies • Sets of triples form a Graph
- 30. Graphs • Triples are building blocks of Graphs • Combining sets of triples allows the construction of arbitrarily complex graphs b4x:martijn b4x:karmeliethas_favorite_beer
- 34. Add meaning ! • Reuse terms from existing, well defined vocabularies – ontologies (foaf, dc, go, so) • Describe new terms = Ontologies • Contain – A crisp human definition – Some machine readable facts
- 35. Metadata • Ontologies are also described in RDF – RDFS: RDF - Schema – OWL: Web Ontology Language – Also expressed in RDF • For clarity, file extension can be .rdfs or .owl
- 36. RDFS Essentials • Descriptions – rdfs:label – rdfs:comment
- 37. RDFS • Relationships between properties, classes – rdfs:Class – rdfs:subClassOf – rdf:Property – rdfs:subPropertyOf – rdfs:range – rdfs:domain
- 38. RDFS: Example @prefix rdfs: <http://www.w3.org/2000/01/rdf-schema#>. @prefix foaf: <http://xmlns.com/foaf/0.1/> . @prefix xsd: <http://www.w3.org/2001/XMLSchema#> . b4x:karmeliet a b4x:Tripel . b4x:Beer a rdfs:Class . b4x:Tripel a rdfs:Class . b4x:Tripel rdfs:subClassOf b4x:Beer . b4x:has_favorite_beer a rdf:Property ; rdfs:domain foaf:Person ; rdfs:range b4x:Beer . b4x:Beer rdfs:subClassOf b4x:Drink .
- 39. Analogy • RDF = database = data • RDFS/OWL = schema = metadata • Both are described in RDF, but have a different scope
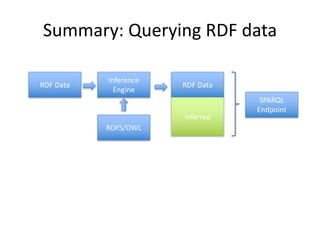
- 40. Semantic Technologies • Inference – Enhance dataset using knowledge from metadata (e.g. rdfs, owl) • Types of inference engines – RDFS inference • RDFS entailment regime – OWL inference • Under active research • Engines exist for specific subsets of OWL (OWL-DL)
- 41. RDFS Entailment
- 42. RDFS: Inference b4x:kevin b4x:has_favorite_beer b4x:stella Q: What can we infer from this using RDFS entailment ?
- 43. RDFS: Inference b4x:kevin b4x:has_favorite_beer b4x:stella Inferred triples: b4x:kevin a foaf:Person [from domain] b4x:stella a b4x:Beer [from range] b4x:stella a b4x:Drink [from subClassOf]
- 44. DuckTyping • Watch out with inference ! Example: You want to express that people can have lengths b4x:length a rdf:Property; rdfs:domain foaf:Person; rdfs:range xsd:integer.
- 45. DuckTyping • Problem: ex:VW_Transporter b4x:length “600”^xsd:integer. • Would infer that VW_Transporter is a Person ! • This is called DuckTyping If it looks like a duck, swims like a duck, and quacks like a duck, then it probably is a duck
- 47. Task • Find a solution: express in rdfs that people can have lengths b4x:havingLenght a rdfs:Class. b4x:length a rdf:Property; rdfs:domain b4x:havingLength; rdfs:range xsd:integer. foaf:Person rdfs:subClassOf b4x:havingLength.
- 48. Storing RDF • As an RDF file for download • In a Triplestore – Database optimised for storing triples – Examples: BlazeGraph, Fuseki, Sesame
- 49. Semantic Technologies • Querying over RDF data: SPARQL • Cool features: – Distributed querying = actual distribution of data and computing resources – SPARQL/Update: modify data • SPARQL endpoints: SPARQL over HTTP
- 50. SPARQL Query Syntax • First example: SELECT ?subject ?predicate ?object WHERE { ?subject ?predicate ?object. } (Generally not a good idea as it will pull down the whole dataset) Binding variables Graph matching
- 51. ? SELECT ?person WHERE { ?person b4x:has_favorite_beer b4x:karmeliet }
- 52. ?
- 53. SPARQL Query Syntax • Limit result size : SELECT ?subject ?predicate ?object WHERE { ?subject ?predicate ?object. } LIMIT 10
- 54. SPARQL Query Syntax • Find all classes: PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#> SELECT ?class ?label WHERE { ?class a rdfs:Class. ?class rdfs:label ?label. } (This will only retrieve classes that have a label)
- 55. SPARQL Query Syntax • Find all classes: PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#> SELECT ?class ?label WHERE { ?class a rdfs:Class. OPTIONAL { ?class rdfs:label ?label. } }
- 56. SPARQL Query Syntax • Find all classes that contain “duck” in the label: PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#> SELECT ?class ?label WHERE { ?class a rdfs:Class. ?class rdfs:label ?label. FILTER( CONTAINS (str(?label) , “duck” ) ) }
- 57. SPARQL Query Syntax • Make it case insensitive: PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#> SELECT ?class ?label WHERE { ?class a rdfs:Class. ?class rdfs:label ?label. FILTER( CONTAINS ( UCASE(str(?label)) , “DUCK” ) ) }
- 58. SPARQL Query Syntax • Search in specific graph: PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#> SELECT ?class ?label FROM <http://example.org/animals> WHERE { ?class a rdfs:Class. ?class rdfs:label ?label. FILTER( CONTAINS ( UCASE(str(?label)) , “DUCK” ) ) }
- 59. SPARQL Query Syntax • Search in specific graph: PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#> SELECT ?class ?label WHERE { GRAPH <http://example.org/animals> { ?class a rdfs:Class. ?class rdfs:label ?label. FILTER( CONTAINS ( UCASE(str(?label)) , “DUCK” ) ) } }
- 60. SPARQL Query Syntax • Can also search for graphs : PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#> SELECT ?g WHERE { GRAPH ?g { ?class a rdfs:Class. ?class rdfs:label ?label. FILTER( CONTAINS ( UCASE(str(?label)) , “DUCK” ) ) } }
- 62. • Basic data element = a Triple – A mini sentence – Contains three Terms: – Subject Predicate Object • Example: <http://xmpl/entities#martijn> <http://xmpl/relations#has_favorite_beer> <http://xmpl/entities#karmeliet>. Take home Summary
- 63. • Combine triples to represent knowledge
- 64. • Use terms from ONTOLOGIES – COMMON VOCABULARIES – POSSIBLE TO INFER MEANING • OMIABIS • OBIB • SNOMED/ICD • MESH
- 65. ? • SPARQL searches for patterns
- 66. ?
- 67. Interoperability between OBO and Semantic Technologies • Originated from two separate academic worlds • Computing applications of OBO mainly consistency checking and overrepresentation analysis • Semantic Technologies: much broader toolset • Interoperability ? – Direct offering in both formats – Automated mappings • Migration towards semantic toolkits
- 68. Where to find ontologies • OBO Foundry • Bioportal; NCBO • Biogateway • Bio2RDF
- 69. Where to find RDF data • Google for SPARQL endpoint • => e.g. EBI databases • Non biological: DBpedia
- 70. How about Tim Berners Lee’s vision • We’re not there yet, but for bio data we’re getting quite close – The explicitome – Crowd sourcing – Nanopublications
- 72. SPARQL : Recap PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#> SELECT ?label FROM <http://graphName> WHERE { ?x rdfs:label ?label. FILTER ( CONTAINS(?label, “dimethylalinine”) ) } LIMIT 10 ORDER BY ?label
- 73. SPARQL : Recap PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#> SELECT ?label FROM <http://graphName> WHERE { ?x rdfs:label ?label. FILTER ( CONTAINS(?label, “dimethylalinine”) ) } LIMIT 10 ORDER BY ?label • FIND the pattern ?x rdfs:label ?label.
- 74. SPARQL : Recap PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#> SELECT ?label FROM <http://graphName> WHERE { ?x rdfs:label ?label. FILTER ( CONTAINS(?label, “dimethylalinine”) ) } LIMIT 10 ORDER BY ?label • FIND the pattern ?x rdfs:label ?label. • BIND variables ?label, ?x
- 75. SPARQL : Recap PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#> SELECT ?label FROM <http://graphName> WHERE { ?x rdfs:label ?label. FILTER ( CONTAINS(?label, “dimethylalinine”) ) } LIMIT 10 ORDER BY ?label • FIND the pattern ?x rdfs:label ?label. • BIND variables ?label, ?x • RETRIEVE variable ?label
- 76. SPARQL : Recap PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#> SELECT ?label FROM <http://graphName> WHERE { ?x rdfs:label ?label. FILTER ( CONTAINS(?label, “dimethylalinine”) ) } LIMIT 10 ORDER BY ?label • FIND the pattern ?x rdfs:label ?label. • BIND variables ?label, ?x • RETRIEVE variable ?label • PREFIX: replace rdfs:label by <http://www.w3.org/2000/01/rdf-schema#label>
- 77. SPARQL : Recap PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#> SELECT ?label FROM <http://graphName> WHERE { ?x rdfs:label ?label. FILTER ( CONTAINS(?label, “dimethylalinine”) ) } LIMIT 10 ORDER BY ?label • FIND the pattern ?x rdfs:label ?label. • BIND variables ?label, ?x • RETRIEVE variable ?label • PREFIX: replace rdfs:label by <http://www.w3.org/2000/01/rdf-schema#> • FILTER results to labels containing “dimethylalinine”
- 78. SPARQL : Recap PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#> SELECT ?label FROM <http://graphName> WHERE { ?x rdfs:label ?label. FILTER ( CONTAINS(?label, “dimethylalinine”) ) } LIMIT 10 ORDER BY ?label • FIND the pattern ?x rdfs:label ?label. • BIND variables ?label, ?x • RETRIEVE variable ?label • PREFIX: replace rdfs:label by <http://www.w3.org/2000/01/rdf-schema#> • FILTER results to labels containing “dimethylalinine” • LIMIT results to first 10 matches ordered by label
- 81. • From a web interface • Using http – HTTP GET – HTTP POST : for larger query strings – Headers determine response type (JSON, XML, HTML) http://…/sparql?default-graph-uri=<http://graphName>&query=URLENCODEDQUERYSTRING Running SPARQL
- 82. BIO-ONTOLOGIES
- 83. BioPortal
- 84. Access • From the web interface ! • SPARQL endpoint: using API key; on request • Running a local copy: download VM image; on request
- 85. Exercises • Find a term • Find ontologies containing a term • Browse some ontologies • Check the NCBO annotator !
- 86. BIO-DATA
- 89. Ensembl
- 92. PREFIX up:<http://purl.uniprot.org/core/> PREFIX taxon:<http://purl.uniprot.org/taxonomy/> PREFIX rdfs:<http://www.w3.org/2000/01/rdf-schema#> PREFIX obo:<http://purl.obolibrary.org/obo/> SELECT * WHERE { ?protein up:classifiedWith obo:GO_0004499. ?protein up:organism taxon:9606. } http://sparql.uniprot.org
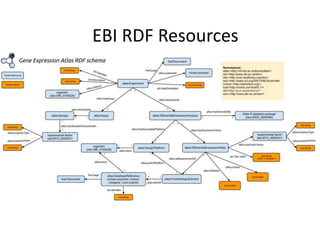
- 94. PREFIX rdf: <http://www.w3.org/1999/02/22-rdf-syntax-ns#> PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#> PREFIX owl: <http://www.w3.org/2002/07/owl#> PREFIX dcterms: <http://purl.org/dc/terms/> PREFIX obo: <http://purl.obolibrary.org/obo/> PREFIX sio: <http://semanticscience.org/resource/> PREFIX efo: <http://www.ebi.ac.uk/efo/> PREFIX atlas: <http://rdf.ebi.ac.uk/resource/atlas/> PREFIX atlasterms: <http://rdf.ebi.ac.uk/terms/atlas/> PREFIX up:<http://purl.uniprot.org/core/> PREFIX biopax3:<http://www.biopax.org/release/biopax-level3.owl#> SELECT distinct ?protein ?expressionValue ?pvalue WHERE { ?factor rdf:type efo:EFO_0000384 . ?value atlasterms:hasFactorValue ?factor . ?value atlasterms:isMeasurementOf ?probe . ?value atlasterms:pValue ?pvalue . ?value rdfs:label ?expressionValue . ?probe atlasterms:dbXref ?protein . FILTER ( ?pvalue < 1e-12 ) FILTER ( strstarts(str(?protein),"http://purl.uniprot.org/uniprot/") ) }ORDER BY ASC (?pvalue) https://www.ebi.ac.uk/rdf/services/atlas/sparql
- 95. • Links pathways with genes, terms from Pathway, Cell line and Disease ontology, PubMed references • Models individual Interactions • Can be downloaded as RDF • Has an experimental SPARQL endpoint WikiPathways
- 97. PREFIX wp: <http://vocabularies.wikipathways.org/wp#> PREFIX dc: <http://purl.org/dc/elements/1.1/> PREFIX dcterms: <http://purl.org/dc/terms/> PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#> SELECT DISTINCT ?PathwayName where { ?geneProduct a wp:GeneProduct . ?geneProduct dc:identifier ?GeneID . ?geneProduct dcterms:isPartOf ?pathway . ?geneProduct rdfs:label ?geneName . ?pathway dc:identifier ?pathwayid . ?pathway dc:title ?PathwayName . FILTER(str(?geneName) = "TNFalpha" ) } http://sparql.wikipathways.org
- 100. • Try this, or another query – Using web interface – Using http get • Define a simple describe • Use a web tool to URLEncode the query • Submit query as a URL parameter Exercise
- 101. DisGeNet
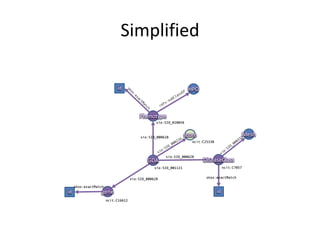
- 105. PREFIX rdf: <http://www.w3.org/1999/02/22-rdf-syntax-ns#> PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#> PREFIX owl: <http://www.w3.org/2002/07/owl#> PREFIX xsd: <http://www.w3.org/2001/XMLSchema#> PREFIX dcterms: <http://purl.org/dc/terms/> PREFIX foaf: <http://xmlns.com/foaf/0.1/> PREFIX skos: <http://www.w3.org/2004/02/skos/core#> PREFIX void: <http://rdfs.org/ns/void#> PREFIX sio: <http://semanticscience.org/resource/> PREFIX ncit: <http://ncicb.nci.nih.gov/xml/owl/EVS/Thesaurus.owl#> PREFIX up: <http://purl.uniprot.org/core/> SELECT DISTINCT ?disease WHERE { ?gda a sio:SIO_000983. ?gda sio:SIO_000628 ?disease. ?disease a ncit:C7057. ?gda sio:SIO_000628 ?gene. ?gene a ncit:C16612. ?gene skos:exactMatch <http://identifiers.org/hgnc.symbol/BRCA1>} http://rdf.disgenet.org/lodestar/sparql
- 107. PREFIX rdf: <http://www.w3.org/1999/02/22-rdf-syntax-ns#> PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#> PREFIX owl: <http://www.w3.org/2002/07/owl#> PREFIX xsd: <http://www.w3.org/2001/XMLSchema#> PREFIX dcterms: <http://purl.org/dc/terms/> PREFIX foaf: <http://xmlns.com/foaf/0.1/> PREFIX skos: <http://www.w3.org/2004/02/skos/core#> PREFIX void: <http://rdfs.org/ns/void#> PREFIX sio: <http://semanticscience.org/resource/> PREFIX ncit: <http://ncicb.nci.nih.gov/xml/owl/EVS/Thesaurus.owl#> PREFIX up: <http://purl.uniprot.org/core/> SELECT DISTINCT ?disease WHERE { ?gda a sio:SIO_000983. ?gda sio:SIO_000628 ?disease. ?disease a ncit:C7057. ?gda sio:SIO_000628 ?gene. ?gene a ncit:C16612. ?gene skos:exactMatch <http://identifiers.org/hgnc.symbol/BRCA1>} http://rdf.disgenet.org/lodestar/sparql
- 108. PREFIX rdf: <http://www.w3.org/1999/02/22-rdf-syntax-ns#> PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#> PREFIX owl: <http://www.w3.org/2002/07/owl#> PREFIX xsd: <http://www.w3.org/2001/XMLSchema#> PREFIX dcterms: <http://purl.org/dc/terms/> PREFIX foaf: <http://xmlns.com/foaf/0.1/> PREFIX skos: <http://www.w3.org/2004/02/skos/core#> PREFIX void: <http://rdfs.org/ns/void#> PREFIX sio: <http://semanticscience.org/resource/> PREFIX ncit: <http://ncicb.nci.nih.gov/xml/owl/EVS/Thesaurus.owl#> PREFIX up: <http://purl.uniprot.org/core/> SELECT DISTINCT ?disease WHERE { ?gda a [(rdfs:subClassOf)* sio:SIO_000983]. ?gda sio:SIO_000628 ?disease. ?disease a ncit:C7057. ?gda sio:SIO_000628 ?gene. ?gene a ncit:C16612. ?gene skos:exactMatch <http://identifiers.org/hgnc.symbol/BRCA1>} http://rdf.disgenet.org/lodestar/sparql
- 111. PREFIX xsd: <http://www.w3.org/2001/XMLSchema#> PREFIX dcterms: <http://purl.org/dc/terms/> PREFIX foaf: <http://xmlns.com/foaf/0.1/> PREFIX skos: <http://www.w3.org/2004/02/skos/core#> PREFIX void: <http://rdfs.org/ns/void#> PREFIX sio: <http://semanticscience.org/resource/> PREFIX ncit: <http://ncicb.nci.nih.gov/xml/owl/EVS/Thesaurus.owl#> PREFIX up: <http://purl.uniprot.org/core/> SELECT DISTINCT ?gene WHERE { ?gda sio:SIO_000628 ?gene,?disease . ?gene a ncit:C16612 . ?gene skos:exactMatch ?GeneID . ?disease a ncit:C7057 . ?disease dcterms:title ?DiseaseName . ?gda sio:SIO_000216 ?scoreIRI . ?scoreIRI sio:SIO_000300 ?score . FILTER (?score > "0.35"^^xsd:decimal) FILTER (contains(str(?DiseaseName),"Crohn")) } http://rdf.disgenet.org/lodestar/sparql
- 113. neXtProt
- 115. select distinct ?id where { ?entry skos:exactMatch ?id. ?entry :isoform ?isoform. ?isoform :medical ?medical_annotation. ?medical_annotation :term ?term. ?term :related ?disease. ?disease a :MeshCv. ?disease rdfs:label ?label. FILTER(CONTAINS(?label,"Cardio")). } https://snorql.nextprot.org/
- 117. select ?chrom ?start ?end where { ?gene rdf:type :Gene. ?gene :name ?name. ?gene :chromosome ?chrom. ?gene :begin ?start. ?gene :end ?end. FILTER (str(?name) = "TP53") } https://snorql.nextprot.org/
- 120. PREFIX rdf: <http://www.w3.org/1999/02/22-rdf-syntax-ns#> PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#> PREFIX owl: <http://www.w3.org/2002/07/owl#> PREFIX xsd: <http://www.w3.org/2001/XMLSchema#> PREFIX dcterms: <http://purl.org/dc/terms/> PREFIX foaf: <http://xmlns.com/foaf/0.1/> PREFIX skos: <http://www.w3.org/2004/02/skos/core#> PREFIX void: <http://rdfs.org/ns/void#> PREFIX sio: <http://semanticscience.org/resource/> PREFIX ncit: <http://ncicb.nci.nih.gov/xml/owl/EVS/Thesaurus.owl#> PREFIX up: <http://purl.uniprot.org/core/> PREFIX wp: <http://vocabularies.wikipathways.org/wp#> PREFIX dc: <http://purl.org/dc/elements/1.1/> PREFIX dcterms: <http://purl.org/dc/terms/> http://rdf.disgenet.org/lodestar
- 121. SELECT DISTINCT ?PathwayName WHERE { ?gda sio:SIO_000628 ?gene, ?disease . ?gene a ncit:C16612 . ?disease a ncit:C7057 . ?disease dcterms:title ?DiseaseName . ?gda sio:SIO_000216 ?scoreIRI . ?scoreIRI sio:SIO_000300 ?score . FILTER (?score > "0.35"^^xsd:decimal) FILTER (contains(str(?DiseaseName),"Crohn")) SERVICE <http://sparql.wikipathways.org/> { ?geneProduct a wp:GeneProduct . ?geneProduct dc:identifier ?gene . ?geneProduct dcterms:isPartOf ?pathway . ?pathway dc:identifier ?pathwayid . ?pathway dc:title ?PathwayName . } } http://rdf.disgenet.org/lodestar/sparql
- 124. Functionalities • Uploader/converter – Upload BED/GFF/VCF files – Automatically translated into triples – Stored in triplestore
- 125. Functionalities • Query builder – Visually build SPARQL queries that query your data along with public data – Store results as new graphs or download as CSV

























![The Turtle Syntax
• Blank nodes
@prefix b4x: <http:bioinformatics.be/terms#> .
@prefix foaf: <http://xmlns.com/foaf/0.1/> .
@prefix xsd: <http://www.w3.org/2001/XMLSchema#> .
b4x:martijn b4x:has_favorite_beer b4x:karmeliet,
b4x:chimay_blauw;
b4x:length “184”^^xsd:integer;
foaf:name “Martijn Devisscher”^^xsd:string;
b4x:owns_cat [ b4x:color “Gray” ].](https://image.slidesharecdn.com/bioontologiesandsemantictechnologies2-180509123734/85/Bio-ontologies-and-semantic-technologies-2-25-320.jpg)

















![RDFS: Inference
b4x:kevin b4x:has_favorite_beer b4x:stella
Inferred triples:
b4x:kevin a foaf:Person [from domain]
b4x:stella a b4x:Beer [from range]
b4x:stella a b4x:Drink [from subClassOf]](https://image.slidesharecdn.com/bioontologiesandsemantictechnologies2-180509123734/85/Bio-ontologies-and-semantic-technologies-2-43-320.jpg)




















































![Bio ontologies and semantic technologies[2]](https://image.slidesharecdn.com/bioontologiesandsemantictechnologies2-180509123734/85/Bio-ontologies-and-semantic-technologies-2-98-320.jpg)
![Bio ontologies and semantic technologies[2]](https://image.slidesharecdn.com/bioontologiesandsemantictechnologies2-180509123734/85/Bio-ontologies-and-semantic-technologies-2-99-320.jpg)


![Bio ontologies and semantic technologies[2]](https://image.slidesharecdn.com/bioontologiesandsemantictechnologies2-180509123734/85/Bio-ontologies-and-semantic-technologies-2-103-320.jpg)




![PREFIX rdf: <http://www.w3.org/1999/02/22-rdf-syntax-ns#>
PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#>
PREFIX owl: <http://www.w3.org/2002/07/owl#>
PREFIX xsd: <http://www.w3.org/2001/XMLSchema#>
PREFIX dcterms: <http://purl.org/dc/terms/>
PREFIX foaf: <http://xmlns.com/foaf/0.1/>
PREFIX skos: <http://www.w3.org/2004/02/skos/core#>
PREFIX void: <http://rdfs.org/ns/void#>
PREFIX sio: <http://semanticscience.org/resource/>
PREFIX ncit: <http://ncicb.nci.nih.gov/xml/owl/EVS/Thesaurus.owl#>
PREFIX up: <http://purl.uniprot.org/core/>
SELECT DISTINCT ?disease WHERE {
?gda a [(rdfs:subClassOf)* sio:SIO_000983].
?gda sio:SIO_000628 ?disease.
?disease a ncit:C7057.
?gda sio:SIO_000628 ?gene.
?gene a ncit:C16612.
?gene skos:exactMatch <http://identifiers.org/hgnc.symbol/BRCA1>}
http://rdf.disgenet.org/lodestar/sparql](https://image.slidesharecdn.com/bioontologiesandsemantictechnologies2-180509123734/85/Bio-ontologies-and-semantic-technologies-2-108-320.jpg)



![Bio ontologies and semantic technologies[2]](https://image.slidesharecdn.com/bioontologiesandsemantictechnologies2-180509123734/85/Bio-ontologies-and-semantic-technologies-2-112-320.jpg)



![Bio ontologies and semantic technologies[2]](https://image.slidesharecdn.com/bioontologiesandsemantictechnologies2-180509123734/85/Bio-ontologies-and-semantic-technologies-2-116-320.jpg)

![Bio ontologies and semantic technologies[2]](https://image.slidesharecdn.com/bioontologiesandsemantictechnologies2-180509123734/85/Bio-ontologies-and-semantic-technologies-2-118-320.jpg)



![Bio ontologies and semantic technologies[2]](https://image.slidesharecdn.com/bioontologiesandsemantictechnologies2-180509123734/85/Bio-ontologies-and-semantic-technologies-2-122-320.jpg)



