Chou fasman algorithm for protein structure prediction
- 2. Contents… • Importance of the Structures of proteins • Prediction of 2D Structures • Chou-Fasman Algorithm • How it works! Chou-Fasman Algorithm for Protein Prediction 2
- 3. What is chou-fasman algorithm? • The experimental methods used by biotechnologists to determine the structures of proteins demand sophisticated equipment and time. • A host of computational methods are developed to predict the location of secondary structure elements in proteins for complementing or creating insights into experimental results. • Chou-Fasman algorithm is an empirical algorithm developed for the prediction of protein secondary structure Chou-Fasman Algorithm for Protein Prediction 3
- 4. Before we go….. • Structures of proteins…… • Why study of structures are important…. • What is the need of an algorithm …. Chou-Fasman Algorithm for Protein Prediction 4
- 5. Chou-Fasman Algorithm for Protein Prediction 5
- 6. Secondary structure prediction • In either case, amino acid propensities should be useful for predicting secondary structure • Two classical methods that use previously determined propensities: • Chou-Fasman • Garnier-Osguthorpe-Robson Chou-Fasman Algorithm for Protein Prediction 6
- 7. Goal… • Take primary structure (sequence) and, using rules derived from known structures, predict the secondary structure that is most likely to be adopted by each residue • Major classes are a-helices, b-sheets and loops Chou-Fasman Algorithm for Protein Prediction 7
- 8. Structural Propensities • Due to the size, shape and charge of its side chain, each amino acid may “fit” better in one type of secondary structure than another • Classic example: The rigidity and side chain angle of proline cannot be accomodated in an a-helical structure Chou-Fasman Algorithm for Protein Prediction 8
- 9. Structural Propensities • Two ways to view the significance of this preference (or propensity) • It may control or affect the folding of the protein in its immediate vicinity (amino acid determines structure) • It may constitute selective pressure to use particular amino acids in regions that must have a particular structure (structure determines amino acid) Chou-Fasman Algorithm for Protein Prediction 9
- 10. Chou-Fasman method • Uses table of conformational parameters (propensities) determined primarily from measurements of secondary structure by CD spectroscopy • Table consists of one “likelihood” for each structure for each amino acid Chou-Fasman Algorithm for Protein Prediction 10
- 11. Chou-Fasman Algorithm for Protein Prediction 11 Chou-Fasman Algorithm • Conformational parameters for every amino acid (AA): P(a) = propensity in an alpha helix P(b) = propensity in a beta sheet P(turn) = propensity in a turn Based on observed propensities in proteins of known structure
- 12. Chou-Fasman propensities (partial table) Amino Acid Pa Pb Pt Glu 1.51 0.37 0.74 Met 1.45 1.05 0.60 Ala 1.42 0.83 0.66 Val 1.06 1.70 0.50 Ile 1.08 1.60 0.50 Tyr 0.69 1.47 1.14 Pro 0.57 0.55 1.52 Gly 0.57 0.75 1.56 Chou-Fasman Algorithm for Protein Prediction 12
- 13. Chou-Fasman method • A prediction is made for each type of structure for each amino acid • Can result in ambiguity if a region has high propensities for both helix and sheet (higher value usually chosen, with exceptions) Chou-Fasman Algorithm for Protein Prediction 13
- 14. Chou-Fasman method • Calculation rules are somewhat ad hoc • Example: Method for helix • Search for nucleating region where 4 out of 6 a.a. have Pa > 1.03 • Extend until 4 consecutive a.a. have an average Pa < 1.00 • If region is at least 6 a.a. long, has an average Pa > 1.03, and average Pa > average Pb consider region to be helix Chou-Fasman Algorithm for Protein Prediction 14
- 15. • Scan the peptide and identify regions where 3 out of 5 contiguous residues have P(β)>100. • These residues nucleate β- strands. Extend these in both directions until a set of four contiguous residues have an average P(β)<100. • This ends β- strand. Chou-Fasman Algorithm for Protein Prediction 15
- 16. • region containing overlapping α and β Any assignment are taken to be helical or β depending on if the average P(α) and P(β) for that region is largest. • If this residues an α or β- region so that it becomes less than 5 residues, the α or β assignment for that region is removed. Chou-Fasman Algorithm for Protein Prediction 16
- 17. Chou-Fasman Algorithm for Protein Prediction 17 SPASEASDGQSVSV P(a) P(b) S: 77 75 P: 55 55 A: 142 83 S: 77 SPASEASDGQFETTY P(a) P(b) E: 151 37 A: 142 83 S: 77 75 D: 101 54 G: 57 Q: 111 1) 4 of 6, P(a) > 100 2) Extend RIGHT until 4 contiguous Residues have P(a) < 100 3) Calculate SP(a) and SP(b). Is SP(a) > SP(b)? (Do Not Include last 4 in sum) Find potential alpha helix: MFCTYYGNNGEHIELMM MFCTYYGNNGEHIELMM
- 18. Accuracy of Chou-Fasman predictions • Sequences whose 3D structures are known are processed so that each residue is “assigned” to a given secondary structure class by looking at the backbone angles • Three classes most often used (helix=H, sheet=E, turn=C) but sometimes use four classes (helix, sheet, turn, loop) Chou-Fasman Algorithm for Protein Prediction 18 Conclusion…..
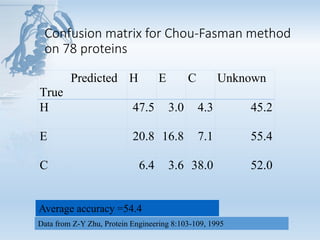
- 19. Confusion matrix for Chou-Fasman method on 78 proteins Predicted True H E C Unknown H 47.5 3.0 4.3 45.2 E 20.8 16.8 7.1 55.4 C 6.4 3.6 38.0 52.0 Data from Z-Y Zhu, Protein Engineering 8:103-109, 1995 Average accuracy =54.4
- 21. Thank You!