How Do You Build and Validate 1500 Models and What Can You Learn from Them?
- 1. © 2018 KNIME AG. All Rights Reserved. How Do You Build and Validate 1500 Models and What Can You Learn from Them? Greg Landrum*, Anna Martin, Daria Goldmann KNIME AG 2018 ICCS @dr_greg_landrum
- 2. © 2018 KNIME AG. All Rights Reserved. The Monster Model Factory Greg Landrum*, Anna Martin, Daria Goldmann KNIME AG 2018 ICCS @dr_greg_landrum
- 3. © 2018 KNIME AG. All Rights Reserved. 3 Who cares? • I have >1500 datasets from ChEMBL that I would like to build models for • I want to actually use the models, so they need to be deployed • The whole process needs to be automated and reproducible so that I can do it again when ChEMBL is updated • Maybe we can learn something interesting from the models themselves
- 4. 4© 2018 KNIME AG. All Rights Reserved. Back to the beginning
- 5. © 2018 KNIME AG. All Rights Reserved. 5 The model process Image from: https://upload.wikimedia.org/wikipedia/commons /b/b9/CRISP-DM_Process_Diagram.png CRISP-DM (CRoss Industry Standard Process for Data Mining) is a standard process for data mining solutions. wikipedia://CRISP-DM
- 6. © 2018 KNIME AG. All Rights Reserved. 6 The model process Image from: https://upload.wikimedia.org/wiki pedia/commons/b/b9/CRISP- DM_Process_Diagram.png Init Load Transform Learn Score Evaluate Deploy
- 7. © 2018 KNIME AG. All Rights Reserved. 7 The model process, multiple models …
- 8. © 2018 KNIME AG. All Rights Reserved. 8 The model process, multiple models …
- 9. © 2018 KNIME AG. All Rights Reserved. 9 The model process, multiple models … https://commons.wikimedia.org/wiki/File:Jabberwocky.jpg
- 10. © 2018 KNIME AG. All Rights Reserved. 10 The model process, multiple models … It’s not feasible to manually do this for a daunting number of models! https://commons.wikimedia.org/wiki/File:Jabberwocky.jpg
- 11. 11© 2018 KNIME AG. All Rights Reserved. https://www.publicdomainpictures.net/view-image.php?image=155188
- 12. © 2018 KNIME AG. All Rights Reserved. 12 Automation: the model process factory
- 13. © 2018 KNIME AG. All Rights Reserved. 13 Init Load Transform Learn Score Evaluate Deploy Automation: the model process factory Score EvaluateTransform DeployLoad Learn Score Learn Load Transform Evaluate Deploy Score EvaluateTransform DeployLoad Learn Score Learn Load Transform Evaluate Deploy Make each step a separate workflow. Use KNIME to orchestrate calling those workflows KNIME blog post: https://goo.gl/LvESqB White paper: https://goo.gl/d6UpUu
- 14. © 2018 KNIME AG. All Rights Reserved. 14 Model Factory Init Load Transform Learn Score Evaluate Deploy
- 15. © 2018 KNIME AG. All Rights Reserved. 15 The heart of the factory: Call Local Workflow1 • Executes another workflow in the same local repository https://pixabay.com/en/heart-veins-arteries-anatomy-152594/ 1 Call Remote Workflow when run on the KNIME Server
- 16. © 2018 KNIME AG. All Rights Reserved. 16 Model Factory Init Load Transform Learn Score Evaluate Deploy
- 17. © 2018 KNIME AG. All Rights Reserved. 17 Model Factory Init Load Transform Learn Score Evaluate Deploy
- 18. 18© 2018 KNIME AG. All Rights Reserved. Details
- 19. © 2018 KNIME AG. All Rights Reserved. 19 Extracting the data • Data source: ChEMBL 23 • Activity types: ('GI50', 'IC50', 'Ki', 'MIC', 'EC50', 'AC50', 'ED50', 'GI', 'Kd', 'CC50', 'LC50', 'MIC90', 'MIC50', 'ID50’) -> 6.5 million points • Define active: Standard_value < 100nM -> 1.3 million actives • Define inactive: Standard_value > 1uM • Define an interesting assay At least 50 actives -> 1556 assays • Final dataset size: 2.5 million data points, 1.5 million compounds Init Load Transform Learn Score Evaluate Deploy
- 20. © 2018 KNIME AG. All Rights Reserved. 20 Init Load Transform Learn Score Evaluate DeployFinding more inactives • The ChEMBL datasets almost all have an unrealistically high ratio of actives to inactives • “Fix” that by adding enough assumed inactives to each dataset to get a 1:10 active:inactive ratio • Pick those assumed inactives to be roughly similar to the actives: Tanimoto similarity of between 0.35 and 0.6 using RDKit Morgan 2 fingerprints
- 21. © 2018 KNIME AG. All Rights Reserved. 21 Extracting the data Init Load Transform Learn Score Evaluate Deploy
- 22. © 2018 KNIME AG. All Rights Reserved. 22 Transform • Convert SMILES from database into chemical structures • Cleanup the chemical structures Init Load Transform Learn Score Evaluate Deploy http://www.rdkit.org
- 23. © 2018 KNIME AG. All Rights Reserved. 23 Transform • Convert SMILES from database into chemical structures • Cleanup the chemical structures • Generate five chemical fingerprints for each structure Init Load Transform Learn Score Evaluate Deploy http://www.rdkit.org
- 24. © 2018 KNIME AG. All Rights Reserved. 24 Transform • Convert SMILES from database into chemical structures • Cleanup the chemical structures • Generate five chemical fingerprints for each structure – Morgan 3 counts (ECFC6), 4K “bits” – Morgan 3 (ECFP6), 4K bits – Morgan 2 (ECFP4), 2K bits – RDKit FP, length 1-5, 2K bits – Atom pairs, distances 1-20, 4K bits Init Load Transform Learn Score Evaluate Deploy http://www.rdkit.org
- 25. © 2018 KNIME AG. All Rights Reserved. 25 Learn and Score Init Load Transform Learn Score Evaluate Deploy 10 different stratified random training/holdout splits generated for each assay
- 26. © 2018 KNIME AG. All Rights Reserved. 26 Learn Init Load Transform Learn Score Evaluate Deploy Learning: • Fingerprint Bayes (NB) • Logistic Regression (LR) • Random Forest (RF) 200 trees, max depth=10, min_leaf_size=3, min_node_size=6 • Gradient Boosting (H2O) 100 trees, max_depth = 5, learning_rate = 0.05 Model Selection: • Pick best model based on Enrichment factor at 5% (EF5)
- 27. © 2018 KNIME AG. All Rights Reserved. 27 Learn Init Load Transform Learn Score Evaluate Deploy Where did these parameters come from? Learning: • Fingerprint Bayes (NB) • Logistic Regression (LR) • Random Forest (RF) 200 trees, max depth=10, min_leaf_size=3, min_node_size=6 • Gradient Boosting (H2O) 100 trees, max_depth = 5, learning_rate = 0.05
- 28. © 2018 KNIME AG. All Rights Reserved. 28 Parameter Optimization Init Load Transform Learn Score Evaluate Deploy • Full parameter optimization done for each method+fingerprint on 70 assays • Results used to pick “standard” parameter sets: – Random Forest: 200 trees, max depth=10, min_leaf_size=3, min_node_size=6 – Gradient Boosting: 100 trees, max_depth = 5, learning_rate = 0.05
- 29. © 2018 KNIME AG. All Rights Reserved. 29 Parameter Optimization Init Load Transform Learn Score Evaluate Deploy
- 30. © 2018 KNIME AG. All Rights Reserved. 30 Parameter Optimization Init Load Transform Learn Score Evaluate Deploy The optimization and model selection workflow is presented in detail in Daria’s KNIME blog post: https://www.knime.com/blog/stuck-in-the-nine-circles-of-hell-try-parameter- optimization-a-cup-of-tea The workflow is available in the EXAMPLES folder inside KNIME: 04_Analytics/11_Optimization/08_Model_Optimization_and_Selection
- 31. 31© 2018 KNIME AG. All Rights Reserved. Making it all run Init Load Transform Learn Score Evaluate Deploy
- 32. © 2018 KNIME AG. All Rights Reserved. 32 Execution • In total >310K models were built1 1 ~1550 assays * 4 methods * 5 FPs * 10 repeats
- 33. © 2018 KNIME AG. All Rights Reserved. 33 Execution KNIME Analytics Platform KNIME Server ... Distributed Executor Distributed Executor Distributed Executor Build/test workflows Run model factory Run individual assays 65-70 load-balanced distributed executors
- 34. 34© 2018 KNIME AG. All Rights Reserved. Are the models any good?
- 35. © 2018 KNIME AG. All Rights Reserved. 35 Performance on validation sets • AUC: mean=0.958 s=0.070 • Cohen’s kappa: mean=0.690 s=0.382
- 36. © 2018 KNIME AG. All Rights Reserved. 36 Performance on validation sets • AUC: mean=0.958 s=0.070 • Cohen’s kappa: mean=0.690 s=0.382 Yeah!
- 37. © 2018 KNIME AG. All Rights Reserved. 37 Performance on validation sets • AUC: mean=0.958 s=0.070 • Cohen’s kappa: mean=0.690 s=0.382 Yeah! Uh oh…
- 38. 38© 2018 KNIME AG. All Rights Reserved. https://www.publicdomainpictures.net/view-image.php?image=155188
- 39. © 2018 KNIME AG. All Rights Reserved. 39 An experiment to check model generalizability • Pick assays where standard_type is Ki • Group them by target ID • Limit to targets where Ki was measured in at least 5 assays -> 11 targets, 73 assays • Use the model built on one assay from a target ID to predict activity across the other assays.
- 40. © 2018 KNIME AG. All Rights Reserved. 40 An experiment to check model generalizability • The targets: TargetID Name Num Assays CHEMBL205 Carbonic anhydrase II 7 CHEMBL224 Serotonin 2a (5-HT2a) receptor 8 CHEMBL234 Dopamine D3 receptor 10 CHEMBL243 Human immunodeficiency virus type 1 protease 6 CHEMBL244 Coagulation factor X 5 CHEMBL253 Cannabinoid CB2 receptor 7 CHEMBL281 Carbonic anhydrase IV 5 CHEMBL3371 Serotonin 6 (5-HT6) receptor 8 CHEMBL344 Melanin-concentrating hormone receptor 1 5 CHEMBL4550 5-lipoxygenase activating protein 5 CHEMBL4908 Trace amine-associated receptor 1 7
- 41. © 2018 KNIME AG. All Rights Reserved. 41 Carbonic Anhydrase IV Carbonic Anhydrase II HIV Protease Factor X 5-HT6 TAAR1
- 42. © 2018 KNIME AG. All Rights Reserved. 42 Carbonic Anhydrase IV Carbonic Anhydrase II HIV Protease Factor X 5-HT6 TAAR1
- 43. © 2018 KNIME AG. All Rights Reserved. 43 An Example Target: CHEMBL3371 (5-HT6) Train on Assay ID: 448716 Test with Assay ID: 1366806 AUROC: 0.38 EF5: 0
- 44. © 2018 KNIME AG. All Rights Reserved. 44 An Example Assay_ID 448716 Assay_ID 1366806
- 45. © 2018 KNIME AG. All Rights Reserved. 45 An Example Target: CHEMBL3371 (5-HT6) Train on Assay ID: 448716 Test with Assay ID: 659849 AUROC: 0.99 EF5: 8.8
- 46. © 2018 KNIME AG. All Rights Reserved. 46 An Example Assay_ID 448716 Assay_ID 659849
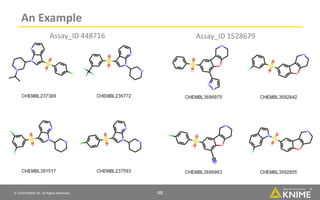
- 47. © 2018 KNIME AG. All Rights Reserved. 47 An Example Target: CHEMBL3371 (5-HT6) Train on Assay ID: 448716 Test with Assay ID: 1528679 AUROC: 0.83 EF5: 0.4
- 48. © 2018 KNIME AG. All Rights Reserved. 48 An Example Assay_ID 448716 Assay_ID 1528679
- 49. © 2018 KNIME AG. All Rights Reserved. 49 Intermediate conclusion • Many/most of the models have likely overfit the training data • Alternative interpretation: we’ve actually built models to predict whether or not a compound is taken from a particular paper • Unfortunately these are functionally the same if you want to predict activity
- 50. 50© 2018 KNIME AG. All Rights Reserved. https://www.publicdomainpictures.net/view-image.php?image=155188
- 51. © 2018 KNIME AG. All Rights Reserved. 51 Look for frequent algorithm + fingerprint combinations • For each of the ~1550 assays * 4 learning algorithms * 10 repeats, look at which fingerprint performed best (as measured by EF5)
- 52. © 2018 KNIME AG. All Rights Reserved. 52 Look for frequent algorithm + fingerprint combinations For each of the ~1550 assays * 4 learning algorithms * 10 repeats, look at which fingerprint performed best (as measured by EF5)
- 53. © 2018 KNIME AG. All Rights Reserved. 53 Which method/FP pair is best for each assay? • For each of the ~1550 assays * 10 repeats, look at which algorithm + fingerprint performed best (as measured by EF51, AUC2, and algorithm complexity3) 1 Rounded to 1 decimal point 2 Rounded to 2 decimal points 3 Random Forest > Gradient Boosting > Fingerprint Bayes > Logistic Regression
- 54. © 2018 KNIME AG. All Rights Reserved. 54 Which method/FP pair is best for each assay? Select best model using EF5, AUC, algorithm complexity
- 55. © 2018 KNIME AG. All Rights Reserved. 55 Wrapping up • We have automated the construction and evaluation of >1500 models for bioassays using data pulled from ChEMBL • We’ve got some strong evidence that the models themselves are significantly overfit • We were able to start to draw some general conclusions about fingerprints and methods
- 56. © 2018 KNIME AG. All Rights Reserved. 56 There’s still a lot left to do • Verify the repeatability of the process by updating when the next version of ChEMBL is released • Some more thought into combining assays to get around the “one series per paper” problem • Look into doing the full optimization run • Come up with a good way of presenting the predictions
- 57. © 2018 KNIME AG. All Rights Reserved. 57 More details… • Model process factory blog post: https://goo.gl/LvESqB • Model process factory white paper: https://goo.gl/d6UpUu • Model process factory workflow: knime://EXAMPLES/50_Applications/26_Model_Process_ Management • Daria’s blog post on the model optimization workflow: https://www.knime.com/blog/stuck-in-the-nine-circles- of-hell-try-parameter-optimization-a-cup-of-tea • Accompanying workflow: knime://EXAMPLES/ 04_Analytics/11_Optimization/08_Model_Optimization_ and_Selection • When we’re done cleaning up, there will be a blog post/sample workflow for the monster model factory too.
- 58. © 2018 KNIME AG. All Rights Reserved. 58 7th RDKit UGM: 19 - 21 September • Hosted by Andreas Bender, Cambridge University • Free registration: https://goo.gl/VVvHUH (or get it on http://www.rdkit.org) http://www.rdkit.org
- 59. © 2018 KNIME AG. All Rights Reserved. 59 KNIME Fall Summit 2018 November 6 – 9 at AT&T Executive Education and Conference Center, Austin, Texas • Tuesday & Wednesday: One-day courses • Thursday & Friday: Summit sessions Use the code ICCS-2018 for 10% off tickets. Register at: knime.com/fall-summit2018
- 60. 60© 2018 KNIME AG. All Rights Reserved. The KNIME® trademark and logo and OPEN FOR INNOVATION® trademark are used by KNIME.com AG under license from KNIME GmbH, and are registered in the United States. KNIME® is also registered in Germany.