Implementation of DNA sequence alignment algorithms using Fpga ,ML,and CNN
- 1. Implementation of DNA/RNA Sequence Alignment Algorithms using FPGA Prepared by :Amr Rashed Under Supervision of : Assoc. Prof. Dr. Hossam El-Din Moustafa Assis. Prof. Dr. Hanan abdelfatah
- 2. Author publications Paper title year journal Impact factor (ISI) Accelerating DNA pairwise sequence alignment using FPGA and a customized convolutional neural network Volume 92, June 2021, 107112 Computers & Electrical Engineering 2.663 Sequence Alignment Using Machine Learning-based Needleman–Wunsch Algorithm Under review IEEE open access
- 3. Aim of the work Highlights Problem Definition Limitations Introduction to bioinformatics S/W Implementation Proposed Fast Technique ( H/W implementation) Convolutional neural network Machine learning 3 AGENDA
- 4. Aim of the work • The proposed implementation relies on the complete parallelization of commonly used sequence alignment algorithms (i.e., the Smith-Waterman algorithm and the Needleman–Wunsch algorithm) under certain limitations using efficient low-cost hardware and software platforms to overcome most of the problems of dynamic programming and hardware implementation.
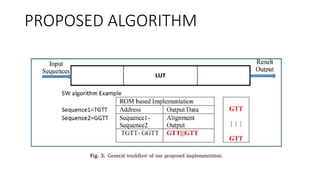
- 5. Highlights • An implementation based on a look-up-table (LUT) to accelerate DNA sequence alignment algorithms under certain limitations is presented. • Our ROM-based hardware implementation requires only O(N/4) cycles or calculation steps to obtain the complete result or a maximum delay of 7.5 ns when implemented using combinational circuits. • The derivation of 254 patterns is presented for a global alignment array for all the input combinations. • It represents a new use of classical ML and deep CNN for global sequence alignment. Fifty-four Boolean functions are derived for complete parallel implementation of the sequence alignment algorithm. • It is valid for RNA/DNA sequences and applicable to software and hardware design. • Hardware implementation can further obtain more tests and be evaluated for long sequences.
- 6. Problem Definition (1) The number of sequences is large, and each of their lengths can be very long. (2) Table II shows that the algorithms used to align the sequences requires O(MN) calculation steps and consumes O(MN) time (M and N are the lengths of the two input sequences). (3) Basic sequence alignment algorithms are internally dependent on the sequential process. (4) Hardware implementation of sequence alignment algorithms does not present an effective solution for sequential process problems, which affects system speed. (5) Practical problems exist owing to parallel implementations, such as the communication overheads. (6) Dynamic algorithms guarantee optimal alignment, but they are slower than FASTA and BLAST, and require extensive computation time and memory because of the sequential processes. Although FASTA and BLAST are fast, they do not guarantee optimum alignment. Our proposed algorithms TABLE II COMPARE SPACE AND TIME COMPLEXITY FOR DP ALGORITHMS [1] Algorithm Type Space Complexity Time Complexity SW Local-linear gap penalty O(MN) O(MN) Gotoh Local-affine gap penalty O(MN) O(MN) Miller– Myers Local-affine gap penalty O(M+N) O(NM) NW Global-linear gap penalty O(MN) O(MN) Hirschberg Global-linear gap penalty O(M+N) O(MN)
- 7. Limitations we propose using equal-length sequences (i.e., multiples of four N=4, 8, 12 ...) that can be applied to DNA or RNA sequences because DNA and RNA sequences consist of four letters of the alphabet, representing four NeocloBases despite the protein sequence which consists of 20 amino acids (letters). The type of alignment used in this study is a pair-wise sequence, and our proposed technique is applied for two algorithms: the SW algorithm for local alignment and the NW algorithm for global alignment. The proposed algorithm can also be applied to other local or global alignment algorithms such as Gotoh, Miller–Myers, and Hirschberg. According to these assumptions, we can implement fully concurrent or parallel software and hardware, which are faster than all the other traditional implementations and do not require extensive computations that are time consuming or power consuming. This implementation is based on a lookup table (LUT). In a special case (NW algorithm), it can be based on a DL model (CNN).
- 8. 8 Introduction to bioinformatics 9 Differences between smith-waterman and Needleman-Wunsch Algorithm 8 Dynamic Programming Algorithm for Sequence Alignment. 7 Public Sequence Databases. 6 Sequence Alignment Types, Description. 5 Dynamic Programming. 4 DNA,RNA Nucleic Acids. 3 Chromosome Structure. 2 From Cell-to-DNA. 1 DEF of Bioinformatics, DNA.
- 9. Bioinformatics Bioinformatics is an interdisciplinary research area at between computer science and biological science. It is a union of biology and informatics as it involves the technology that uses computers for (1)Storage, (2)Retrieval, (3)Manipulation and distribution of information related to biological macromolecules such as DNA, RNA and proteins. Major research efforts includes: (1)Sequence alignment, (2)Gene finding, (3)Genome assembly, drug design, drug discovery , protein structure alignment, protein structure prediction, genome-wise association studies and modeling of association.
- 10. DNA DEFINITION DNA is the hereditary material or complex molecule founds inside every cell in all living things. It contains the instructions an organism needs to develop, live and reproduce. These instructions tell the cell what role it will play in our body. Nearly every cell in a person’s body has the same DNA. Most DNA is located in : (1)The cell nucleus (where it is called nuclear DNA), (2) A small amount of DNA can also be found in the mitochondria (where it is called mitochondrial DNA or mtDNA). Because the cell is very small, and because organisms have many DNA molecules per cell, each DNA molecule must be tightly packaged. This packaged form of the DNA is called a chromosome. An organism's complete set of nuclear DNA is called its genome (our genome is made of a chemical called DNA).
- 12. Chromosome Structure • DNA forms the inherited genetic material inside each cell of a living organism . Each segment in DNA which encodes for a protein is called a gene. 12
- 15. DNA to RNA to Protein, Illustrating the Genetic Code
- 16. gatcctccat atacaacggt atctccacct caggtttaga tctcaacaac ggaaccattg ccgacatgag acagttaggt atcgtcgaga gttacaagct aaaacgagca gtagtcagct ctgcatctga agccgctgaa gttctactaa gggtggataa catcatccgt gcaagaccaa gaaccgccaa tagacaacat atgtaacata tttaggatat acctcgaaaa taataaaccg ccacactgtc attattataa ttagaaacag aacgcaaaaa ttatccacta tataattcaa agacgcgaaa aaaaaagaac aacgcgtcat agaacttttg gcaattcgcg tcacaaataa attttggcaa cttatgtttc ctcttcgagc agtactcgag ccctgtctca agaatgtaat aatacccatc gtaggtatgg ttaaagatag catctccaca acctcaaagc tccttgccga gagtcgccct cctttgtcga gtaattttca cttttcatat gagaacttat tttcttattc tttactctca catcctgtag tgattgacac tgcaacagcc accatcacta gaagaacaga acaattactt aatagaaaaa ttatatcttc ctcgaaacga tttcctgctt ccaacatcta cgtatatcaa gaagcattca cttaccatga cacagcttca gatttcatta ttgctgacag ctactatatc actactccat ctagtagtgg ccacgcccta tgaggcatat cctatcggaa aacaataccc cccagtggca agagtcaatg aatcgtttac atttcaaatt tccaatgata cctataaatc gtctgtagac aagacagctc aaataacata caattgcttc gacttaccga gctggctttc gtttgactct agttctagaa cgttctcagg tgaaccttct tctgacttac tatctgatgc gaacaccacg ttgtatttca atgtaatact cgagggtacg gactctgccg http://www.ncbi.nlm.nih.gov/genbank/samplerecord/ Sample DNA Sequence
- 18. Biological Sequence Alignment • Is a way of arranging two (Pairwise Alignment) or more (Multiple Sequence Alignment) biological sequences (e.g., DNA, RNA, or Protein sequences) of characters to identify regions of similarity. Similarities may be a consequence of functional or evolutionary relationships between these sequences. • Main types : • 1- Pairwise Sequence Alignment. • 2- Multiple Sequence Alignment.
- 19. Pairwise Sequence Alignment Pairwise Alignment Heuristic programming BLAST (local) FASTA (Local) SIM2 Dynamic programming Global Alignment Needleman-Wunsch (NW) Hirschberg Local Alignment Smith-Waterman (SW) Gotoh Miller-Myers Dot-matrix technique (Global)
- 20. Pairwise Sequence Alignment COMPARISON BETWEEN PWSA ALGORITHMS Type Optimal or Exact Methods Suboptimal Methods Optimized Methods Methods SW Local alignment , NW Global alignment Heuristic programming BLAST, FASTA, SIM2 Gotoh, Miller–Myers local alignment, Hirschberg Global alignment Advantages /disadvantages Accurate but not fast Fast but not accurate Accurate and optimized than SW, NW
- 21. Alignment Types Alignment Types Algorithm Examples Exhaustive Alignment Brute Force generates the list of all possible alignments between two sequences, score them and select the alignment with the best score. It is practically useless. Global Alignment Compares the entire sequence of two genomes, end-to- end.This alignment is useful when comparing closely related sequences .Examples:Needleman-Wunsch, Hirschberg. Semi-Global (Glocal)Alignment Searching for the best alignment between a short a long. semi-global alignment is useful is when one sequence is short and the other is very long. Local Alignment Does not look for total sequence but it compares segments of all possible lengths and optimizes the similarity measures. More flexible than the global alignment. Examples :Smith-Waterman, Gotoh, Miller- Myers. Database Search BLAST, FASTA.
- 22. Dynamic Programming •“Divide-and Conquer” strategy •Breaks the problem down into smaller sub-problems 1.Solve the smaller sub-problems optimally. 2.Use the sub-problem solutions to construct the optimal solution to the original problem. •Can be applied to problems that consist of overlapping sub- problems. 22
- 23. Public Sequence Databases •NCBI Gene Bank •http://ncbi.nih.gov •contains many sub-databases •Protein Data Bank •http://www.rcsb.org •contains protein structures •SwissProt •http://www.expasy.org/sprot/ •contains annotated protein sequences •Prosite •http://kr.expasy.org/prosite •contains motifs of protein active sites 23
- 24. Dynamic Programming Algorithm for Sequence Alignment, Steps Process steps Set the reference sequence across the top of the NxM matrix and the read sequence along the side. 1 Initialize the first row and first column of the score matrix (values depend on algorithm) 2 For each element, derive scores from neighboring above, above-left, and left units. 3 For each element, compute match and mismatch scores on above-left score and gap score on above and left scores. Choose maximum of computed score as final score. 4 Once all elements in matrix are filled, find the highest score, which is where the last base in the alignment occurs. 5
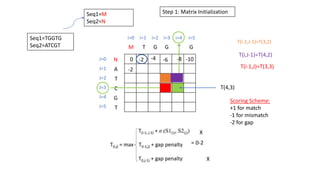
- 29. Seq1=TGGTG Seq2=ATCGT Seq1=M Seq2=N Step 1: Matrix Initialization M T G G T G N A T C G T i=0 i=1 i=2 i=3 i=4 i=5 J=0 J=1 J=2 J=3 J=4 J=5 T(4,3) T(i-1,J-1)=T(3,2) T(i,J-1)=T(4,2) T(i-1,J)=T(3,3) 0 Scoring Scheme: +1 for match -1 for mismatch -2 for gap X X = 0-2 -2 -4 -6 -8 -10 -2
- 33. General workflow
- 34. Smith- waterman Algorithm Example Create Matrix Calculate Matrix Reverse Sequences Backtrack Others
- 35. Comparison between Smith-Waterman Algorithm and the Needleman-Wunch Algorithm 35 Smith-Waterman algorithm Needleman–Wunsch algorithm Type ,Complexity, and Running time Local alignment algorithm, Complexity is O (n2). Runs in O(mn) time, where m and n are the lengths of the two sequences Global alignment algorithm, Complexity is O (n2). Runs in O (mn) time, where m and n are the lengths of the two sequences . Partial or Global Comparison It does not look for the total sequence, but it compares segments of all possible lengths and optimizes the similarity measures It compares the entire sequence of two genomes, end-to-end. Flexibility More Less Suitable for Pairwise alignment Pairwise alignment, similar length, with a significant degree of similarity throughout Gaps It does not penalize gaps in the beginning and end of a sequence. Penalize gaps in the beginning and end of a sequence. Steps: Initialization, scoring Traceback The first row and first column are set to 0. A negative score is set to 0. Begin with the highest score, end when 0 is encountered author shows algorithm steps ، equations ، and scoring matrix. The first row and first column are subject to the gap penalty. The score can be negative. Begin with the cell at the lower right of the matrix, end at the top-left cell Example Sequence 1: GCCCTAGCG Sequence 2: GCGCAATG a match score of +1, a mismatch of -1, and a constant gap penalty of -1 Sequence 1: GCCCTAGCG Sequence 2: GCGCAATG a match score of +1, a mismatch of -1, and a constant gap penalty of -1 GCCCTAGCG | | | GCGCAATG GCCCTAGCG | | : | | : : | GCGC – AATG
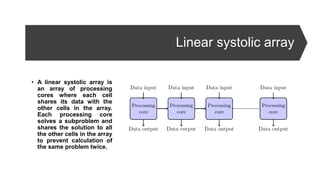
- 36. Linear systolic array • A linear systolic array is an array of processing cores where each cell shares its data with the other cells in the array. Each processing core solves a subproblem and shares the solution to all the other cells in the array to prevent calculation of the same problem twice.
- 39. Binary Representation of DNA Nucleotides
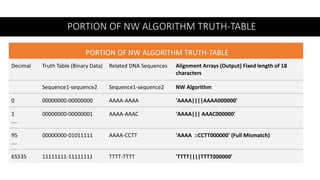
- 40. PORTION OF NW ALGORITHM TRUTH-TABLE PORTION OF NW ALGORITHM TRUTH-TABLE Alignment Arrays (Output) Fixed length of 18 characters Related DNA Sequences Truth Table (Binary Data) Decimal NW Algorithm Sequence1-sequence2 Sequence1-sequence2 'AAAA||||AAAA000000' AAAA-AAAA 00000000-00000000 0 'AAAA||| AAAC000000' AAAA-AAAC 00000000-00000001 1 …. 'AAAA ::CCTT000000' (Full Mismatch) AAAA-CCTT 00000000-01011111 95 …. 'TTTT||||TTTT000000' TTTT-TTTT 11111111-11111111 65535
- 41. PORTION OF SW ALGORITHM TRUTH-TABLE PORTION OF SW ALGORITHM TRUTH-TABLE Alignment Arrays (Output) Fixed length of 12 characters Related DNA Sequences Truth Table (Binary Data) Decimal SW Algorithm Sequence1- Sequence 2 Sequence1-sequence2 'AAAA||||AAAA' AAAA-AAAA 00000000-00000000 0 'AAA|||AAA000' AAAA-AAAC 00000000-00000001 1 …. '000000000000' (Full Mismatch) AAAA-CCTT 00000000-01011111 95 …. 'TTTT||||TTTT' TTTT-TTTT 11111111-11111111 65535
- 42. Relation between length of array and number of instances
- 43. Relation between length of array and number of instances Relationship between Length of Local Alignment Arrays and Number of Instances of Each Length
- 44. Mismatch Conditions DIFFERENCE BETWEEN LOCAL AND GLOBAL ALIGNMENT ARRAYS OF SOME FULL MISMATCH CONDITIONS Local Alignment array (Full Mismatch 12 character) Global Alignment array (NCBI) Global Alignment array (18 character) (Matlab Function) Related DNA Sequences Truth Table (Binary Data) Decimal value Smith-Waterman Algorithm Needleman-Wunsch Algorithm Needleman-Wunsch Algorithm( Full Mismatch) Sequence 1- Sequence 2 Sequence 1- sequence 2 '000000000000' 'AAAA CCCC' 'AAAA CCCC000000' AAAA-CCCC 00000000- 01010101 85 '000000000000' 'AAAA CCCG' 'AAAA :CCCG000000' AAAA-CCCG 00000000- 01010110 86 '000000000000' 'AAAA CCTG' 'AAAA ::CCTG000000' AAAA-CCTT 00000000- 01011111 95 '000000000000' 'AAAA CGGG' 'AAAA :::CGGG000000' AAAA-CGGG 00000000- 01101010 106
- 45. LUT COMPUTATIONAL PARAMETER LUT COMPUTATIONAL PARAMETER Parameters Settings Alignment Algorithm Local, and Global Type of Gap Penalty Linear Gap Opening Gap Opening -5 Gap Extension Gap Extension -5 Substitution Matrix BLOSUM 50 Word Length for each sequence 8-bits (default)
- 46. Analysis of the SW and NW Alignment Arrays Number of Unique and Repeated Alignment Arrays for SW and NW Algorithms NW Algorithm Count (%) SW Algorithm Count (%) Number of Alignment Arrays 65536 (100%) 4836 (7.37%) Unique Alignment arrays 0 (0%) 60700 (92.62%) Remaining Alignment arrays 65536 (100%) 65536 (100%) Total Number
- 47. Analysis of the SW Alignment Arrays Frequent Alignment Arrays for SW Algorithm Percentage Count Frequent Alignment Arrays 6.86% 4498 A|A000000000 12.39% 8124 C|C000000000 9.99% 6548 G|G000000000 6.18% 4051 T|T000000000 35.43% 23221 out of 65536 Total
- 48. Analysis of the SW and NW Alignment Arrays ALIGNMENT ARRAYS ACCORDING TO NUMBER OF MATCHES Type of Alignment Array SW Alignment Array Count NW Alignment Array Count Full mismatch alignment arrays 1812 (2.76%) 9118 (13.91%) Alignment arrays with one match 29371 (44.81%) 24192 (36.91%) Alignment arrays with two matches 28310 (43.19%) 24654 (37.62%) Alignment arrays with three matches 5787 (8.83%) 7316 (11.16%) Full match alignment arrays 256 (0.39%) 256 (0.39%) Total number of alignment arrays (216) 65536 (100%) 65536 (100%)
- 49. Software Performance Evaluation(SW) SW ALGORITHM SOFTWARE IMPLEMENTATION RESULTS FOR SINGLE-CORE AND SIX-CORE PLATFORMS Platform Query Length 4 5000 10k 1M 2M 3M 4M 4.4M Single core Time(s) 30.7µs 0.0360 0.0704 6.8336 13.0377 20.7643 27.2987 30.21 GCUPS 5.6153e- 04 0.645 1.4198 146.3361 290.03 433.43 586.109 640 6-cores Time(s) 45.152 µs 0.0828 0.0821 2.0798 4.0322 5.9160 7.6441 7.8607 GCUPS 3.5436e- 04 0.3019 1.2174 480.8071 992.0214 1521.3 2093.1 2462.9
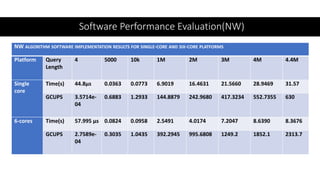
- 50. Software Performance Evaluation(NW) NW ALGORITHM SOFTWARE IMPLEMENTATION RESULTS FOR SINGLE-CORE AND SIX-CORE PLATFORMS Platform Query Length 4 5000 10k 1M 2M 3M 4M 4.4M Single core Time(s) 44.8µs 0.0363 0.0773 6.9019 16.4631 21.5660 28.9469 31.57 GCUPS 3.5714e- 04 0.6883 1.2933 144.8879 242.9680 417.3234 552.7355 630 6-cores Time(s) 57.995 µs 0.0824 0.0958 2.5491 4.0174 7.2047 8.6390 8.3676 GCUPS 2.7589e- 04 0.3035 1.0435 392.2945 995.6808 1249.2 1852.1 2313.7
- 51. Performance comparison with other state-of-the-art implementations Performance comparison with other state-of-the-art implementations Paper Year Platform Sequence Pairs Time (s) GCUPS [15] 2014 1 Xeon Phi D4.4 vs D4.6M 700 29.2 [15] 2014 2 Xeon Phis D4.4 vs D4.6M 396 51.7 [15] 2014 4 Xeon Phis D4.4 vs D4.6M 203 100.7 [16] 2014 Intel® Core™ i7-3770 CPU @ 3.40GHz×8. 256NT vs 265NT 0.317 -- [17] 2019 2×Xeon Gold 6138 Max Query length=5478 -- 734 Ours (SW algorithm) Intel Core I7-9750H 6-cores 2.60 GHz CPUs Same sequence pairs in [21], after cropping the second sequence to 4.4M 7.8607 2462.9 Ours (SW algorithm) Intel Core I7-9750H 6-cores 2.60 GHz CPUs 256NT vs 265NT same length as in [15] 0.1745 -- Ours (NW algorithm) Intel Core I7-9750H 6-cores 2.60 GHz CPUs Same sequence pairs in [21], after cropping the second sequence to 4.4M 8.3676 2313.7
- 52. Hardware Implementation of SW Algorithm Encode Alignment's Arrays Convert characters into Hexadecimal SW Algorithm Hardware implementation With virtex6 FPGA ROM (with & without clock) Two Design Comparison Flowchart of local alignment hardware implementation
- 53. Encode Alignment's Arrays of Smith- Waterman Algorithm CHARACTERS AND PROPOSED HEX-DECIMAL REPRESENTATION Decimal Binary Hex- Decimal Function Description Character 0 0000 X’0’ Padding Zero ‘0’ 1 0001 X’1’ Match Vertical bar ‘|’ 2 0010 X’2’ Mismatch Colon ‘:’ 3 0011 X’3’ Gap Hyphen ‘-‘ 4 0101 X’4’ Mismatch Space ‘ ‘ 10 1010 X’A’ Nucleotide A ‘A’ 11 1011 X’B’ Nucleotide C ‘C’ 12 1100 X’C’ Nucleotide G ‘G’ 13 1101 X’D’ Nucleotide T ‘T’
- 54. Examples EXAMPLES OF LOCAL ALIGNMENT ARRAYS AND ASSOCIATED HEX-DECIMAL REPRESENTATION DNA INPUT SEQUENCES DECIMAL VALUE ALIGNMENT ARRAYS HEXADECIMAL REPRESENTATION ‘AAAA-AAAA’ 0 ‘AAAA||||AAAA’ ‘AAAA1111AAAA’ ‘AAAA-AAAC’ 1 ‘AAA|||AAA000’ ‘AAA111AAA000’ ‘AAAA-AAAG’ 2 ‘AAA|||AAA000’ ‘AAA111AAA000’ ‘AAAA-AAAT’ 3 ‘AAA|||AAA000’ ‘AAA111AAA000’ ‘AAAA-AACA’ 4 ‘AAAA|| |AACA’ ‘AAAA1141AABA’
- 57. Simulation results Simulation Result of 65536 ×48 ROM using Altera Quartus Prime Pro 16.0
- 58. System Design Summary using FPGA Virtex 6 Table XVII System Design Summary using Virtex 6- XC6VLX240T-1FF1156 Design Name/Basic Feature Number Using Block RAM Only Number of Slices LUT Number of LUT flip flop pairs used Maximum Delay(ns) or Max frequency (MHZ) Estimated Power using Xpower Design 1 (with clock) 96/416(2 3%) -- -- 400MHZ,2.499(ns) 3.422 W Design 2(without clock) -- 95500/150720(63%) 95500(100%) 65MHZ,15.355(ns) 3.422 W
- 59. Hardware Implementation of NW Algorithm Comparison between three CNN designs NW Algorithm Encode alignment's arrays Convert characters to binary Class Reduction 1- Replace amino acid letters by asterisk '*' (254 classes) 2- Merge all full mismatched patterns into one pattern (239 classes) Test classical ML classifiers with four datasets Reshape input sequences to 2D input matrix Implementation of 2D-CNN Boolean function minimizations Hardware implementation using Xilinx FPGA combinational circuits Comparison between four different designs Block diagram of the software and hardware implementation of global alignment algorithm
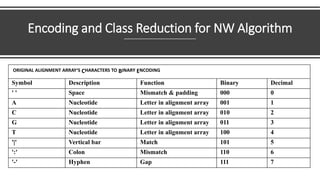
- 60. Encoding and Class Reduction for NW Algorithm ORIGINAL ALIGNMENT ARRAY'S CHARACTERS TO BINARY ENCODING Symbol Description Function Binary Decimal ' ' Space Mismatch & padding 000 0 A Nucleotide Letter in alignment array 001 1 C Nucleotide Letter in alignment array 010 2 G Nucleotide Letter in alignment array 011 3 T Nucleotide Letter in alignment array 100 4 '|' Vertical bar Match 101 5 ':' Colon Mismatch 110 6 '-' Hyphen Gap 111 7
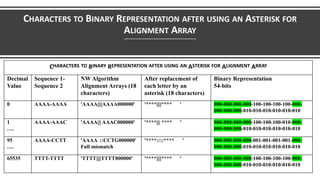
- 61. CHARACTERS TO BINARY REPRESENTATION AFTER USING AN ASTERISK CHARACTERS TO BINARY REPRESENTATION AFTER USING AN ASTERISK Symbol Description Function Binary Decimal '*' Asterisk Represent letters in alignment array 000 0 ':' Colon Mismatch 001 1 ' ' Space Mismatch & padding 010 2 '-' Hyphen Gap 011 3 '|' Vertical bar Match 100 4
- 62. COMPARISON BETWEEN WIDELY USED LOGIC FUNCTION MINIMIZATION METHODS COMPARISON BETWEEN WIDELY USED LOGIC FUNCTION MINIMIZATION METHODS Karnaugh Map (K-Map) Quine–McCluskey (QM) Espresso algorithm Definition Method for simplifying Boolean algebra expressions Known as the tabulation method or the technique of prime implicants, it is functionally similar to Karnaugh Maps. A radically different approach, the algorithm manipulates "cubes," performing the product terms in the ON-, DC-, and OFF covers iteratively. Features Four variables. Unsuitable for more than 6 input variables Tedious and error-prone process It can be performed manually and does not support more than 8 input bits. Challenge to be implemented in computer programs[66][67]. K-Map is not suitable for our algorithms because these have 16 input bits. Can still be performed manually on paper Scales to many variables (Can handle up to 40 variables) One of the highly effective techniques for simplifying Boolean expressions. More convenient to be implemented in computer programs. Has a tabular form that makes it more efficient for use in computer algorithms Has a settled methodology to test whether the minimal form of a Boolean function has been attained. For a larger number of input variables, QM is more effective in minimizing logic functions than K-Map[66][67]. Is not guaranteed to be the global minimum. Practically, it is very closely approximated and free from redundancy. Computationally efficient in terms of both memory requirement and time than the other methods (K-Map and QM) by several orders of magnitude Used as a standard logic expression minimization in logic synthesis tools
- 63. EVOLUTION OF THE NUMBER OF MINTERMS IN EACH BOOLEAN FUNCTION EVOLUTION OF THE NUMBER OF MINTERMS IN EACH BOOLEAN FUNCTION Functions Original "A,C,G,T" minterms count First Reduction (254) minterms count Second Reduction (239) minterms count Fast Minimization minterms count Exact Minimization minterms count F0 23627 0 0 0 0 F1 35562 8590 8590 782 763 F2 37410 8590 8590 782 763 F3 17854 0 0 0 0 F4 34350 2347 2347 467 467 … F52 616 65102 65102 150 150 F53 732 452 452 176 176 TOTAL 1101952 701012 701012 29858 26538
- 64. NW ALIGNMENT STATISTICS NW ALIGNMENT STATISTICS NW Alignment arrays after Replacement of each letter by an asterisk Count Alignment Arrays 254 Number of Unique 65282 Number of Repeated 65536 Total Number of Alignment
- 65. CHARACTERS TO BINARY REPRESENTATION AFTER USING AN ASTERISK FOR ALIGNMENT ARRAY CHARACTERS TO BINARY REPRESENTATION AFTER USING AN ASTERISK FOR ALIGNMENT ARRAY Binary Representation 54-bits After replacement of each letter by an asterisk (18 characters) NW Algorithm Alignment Arrays (18 characters) Sequence 1- Sequence 2 Decimal Value 000-000-000-000-100-100-100-100-000- 000-000-000-010-010-010-010-010-010 '****||||**** ' 'AAAA||||AAAA000000' AAAA-AAAA 0 000-000-000-000-100-100-100-010-000- 000-000-000-010-010-010-010-010-010 '****||| **** ' 'AAAA||| AAAC000000' AAAA-AAAC 1 …. 000-000-000-000-001-001-001-001-000- 000-000-000-010-010-010-010-010-010 '****::::**** ' 'AAAA ::CCTG000000' Full mismatch AAAA-CCTT 95 …. 000-000-000-000-100-100-100-100-000- 000-000-000-010-010-010-010-010-010 '****||||**** ' 'TTTT||||TTTT000000' TTTT-TTTT 65535
- 66. Block diagram for Boolean function minimization Truth Table of NW Algorithm Functions Derivation (54 functions) Functions Minimization (fast method, exact method) MATLAB to VHDL syntax conversion Block diagram for Boolean function minimization Check
- 67. Example
- 68. Example Figure Example of F0=0 and F1 (782 minterms) Minterm Boolean functions after fast minimization
- 69. A portion of minimized F1 Boolean function after altering syntax to MATLAB syntax A portion of minimized F1 Boolean function after altering syntax to MATLAB syntax
- 70. A portion of minimized F1 Boolean function after changing the syntax to VHDL syntax
- 71. Simulation results System Behavioral Simulation (ISim) using ISE Design suite 14.7
- 72. System design System Design Summary Device Family Virtex 6 Device Name XC6VLX240T-1FF1156 Analysis and synthesis resource usage summary Slice Logic Utilization Design Name/Basic Feature Design 1 (No Signals or Variables) Design 2 (Signals inside Process) Design 3 (Variables Used) Design 4 (Signals used without process) Number of Slice LUTs 21658/150720 (14%) 21835/150720 (14%) 21592/150720 (14%) 21872/150720 (14%) Number used as Logic 21658/150720 (14%) 21835/150720 (14%) 21592/150720 (14%) 21872/150720 (14%) IO Utilization Number of bonded IOBs 70/600 (11%) 70/600 (11%) 70/600 (11%) 70/600 (11%) Timing Summary Maximum Combinational Path Delay 8.047 ns 7.904 ns 7.731 ns 7.511 ns Power Analysis Estimated Power 3.422 W 3.422 W 3.422 W 3.422 W
- 73. System design summary SYSTEM DESIGN SUMMARY USING VIRTEX 6- XC6VLX240T-1FF1156 Design Name/Basic Feature Number of Slices LUT Maximum Combinational Path Delay (ns) Estimated Power Using Xpower Design 1 (No Signals or Variables) 21658/150720 (14%) 8.047 ns 3.422 W Design 2 (Signals inside Process) 21835 /150720 (14%) 7.904 ns 3.422 W Design 3 (Variables Used) 21592 /150720 (14%) 7.731 ns 3.422 W Design 4 (Signals used without process) 21872 /150720 (14%) 7.511 ns 3.422 W
- 74. Performance evaluation(1) COMPARISON OF THE PERFORMANCE OF VARIED SINGLE-DEVICE IMPLEMENTATIONS OF THE SW/NW ALGORITHM Paper Year Algorithm Circuit Type Technique Device Frequency (MHz) Time (ns) GCUPS [21] 2009 SW/NW Sequential Systolic cell Xilinx Virtex-4 FX100 100 -- 25.6 [22] 2014 SW Sequential Systolic cell Altera Stratix IV EP4SGX230 57.9 17.27 3.71 [23] 2016 SW Sequential Systolic cell Xilinx XC3S1600E 98.7 10.13 23.79 [24] 2017 SW Sequential Systolic cell -- 250 -- -- [24] 2017 SW Sequential Systolic cell -- 250 -- -- Ours -- SW -Design1 Sequential LUT Xilinx Virtex 6 XC6VLX240T 400 2.499 25.6102 Ours -- NW -Design 4 Combinational LUT Xilinx Virtex 6 XC6VLX240T 133 7.511 8.5333
- 75. Performance evaluation(2) COMPARISON OF USAGE OF FPGA RESOURCES Paper Year Device Frequency (MHz) LUT BRAM Others [25] 2017 -- 250 3056/ 537600 (0.57%) 94/3456 (2.72%) FF- 2468 (0.23%) [26] 2018 Stratix IV EP4SGX230KF40C2 125 70,839 / 182,400 (39%) 1,834,562 / 14,625,792 (13%) Total Registers 73,850 [27] 2018 Kintex7 XC7K480T FFG901-3 100 17462 (8%) -- FF-1402 (1%) Latches-1483 (1%) LUT-FF pairs 216 (5%) Ours -- Xilinx Virtex 6 XC6VLX240T 400 0% 96/416 (23%) 0% Ours -- Xilinx Virtex 6 XC6VLX240T -- 21872 /150720 (14%) 0% 0%
- 76. Performance evaluation(3) COMPARISON OF NUMBER OF OPERATIONS, STEPS, OR CLOCK CYCLES FOR COMPLETE ALIGNMENT Paper Year Algorithm Technique # of Operations for complete alignment [9] 2012 SW Systolic array (N+M−1+Trace-back operations) [28] 2018 SW Systolic array (N+M−1+Trace-back operations) [29] 2020 SW Systolic array (N+M−1+Trace-back operations) Ours -- SW/NW LUT (N/4)
- 77. Performance evaluation(4) PERFORMANCE EVALUATION OF SOFTWARE AND HARDWARE IMPLEMENTATION Sequence Pairs Algorithm Type Time 4NT vs 4NT SW Software-single core 30.7 µs 4NT vs 4NT NW Software-single core 44.8 µs 4NT vs 4NT SW Hardware-FPGA-Design 1 2.499 (ns) 4NT vs 4NT NW Hardware-FPGA-Design 4 7.511 (ns)
- 78. CNN for GLOBAL SEQUENCE ALIGNMENT CONVOLUTIONAL NEURAL NETWORK
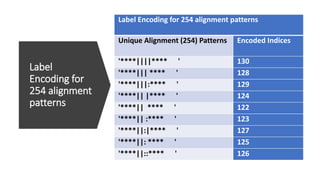
- 79. Label Encoding for 254 alignment patterns Label Encoding for 254 alignment patterns Encoded Indices Unique Alignment (254) Patterns 130 '****||||**** ' 128 '****||| **** ' 129 '****|||:**** ' 124 '****|| |**** ' 122 '****|| **** ' 123 '****|| :**** ' 127 '****||:|**** ' 125 '****||: **** ' 126 '****||::**** '
- 80. A Frequency Table for Alignment 239 Patterns A Frequency Table for Alignment Patterns in Descending Order Percentage Count Unique Alignment Patterns (239 Patterns) 13.9130 % 9118 '****::::**** ' 1.7578 % 1152 '**** | **** ' 1.7578 % 1152 '**** | **** ' 1.5228 % 998 '**** || **** ' 1.4038 % 920 '****| | **** ' 1.4038 % 920 '**** | |**** ' 1.3824 % 906 '**** |: **** ' 1.3824 % 906 '**** :| **** ' 1.2970 % 850 '**** | :**** ' 1.2970 % 850 '****: | **** '
- 81. DATASET DESCRIPTION DATASET DESCRIPTION Database 1 Database 2 Database 3 Reshape of Input Data 16-bit input are reshaped as 4 × 4 matrix Divide input bits into two rows of 8 bits, and zero pad other bits to complete the 8 × 8 matrix. Divide input bits into two rows of 8 bits. Then, repeat each sequence four times to complete the 8 × 8 matrix Number of images 65536 65536 65536 Number of labels 254 254 254
- 82. TRAINING HYPERPARAMETERS TRAINING HYPERPARAMETERS Parameter Value Parameter Value Programming language MATLAB 2020 a Data splitting (train/test) 80/20, randomized Optimizer ADAM, SGDM, RMSPROP Gradient Threshold Inf Maximum Iterations 12270 Max Epochs 30 Learn Rate Schedule Constant Mini Batch Size 128 Learn Rate Drop Factor 0.1000 Execution Environment Single GPU Learn Rate Drop Period 10 Shuffle Every-epoch L2Regularization 10-4 Momentum: 0.9000 Gradient Threshold Method 'l2norm' Initial Learn Rate 0.0100
- 83. Block diagram of customized CNN models Classification Output crossentropyex Softmax Fully Connected fully connected layer with 254 output classes Dropout 50% dropout Fully Connected fully connected layer with 500 output classes ReLU Batch Normalization Convolution 5 filters of size 5x5 convolutions with stride 1 and padding 3 Image Input 8x8x1 or 4x4x1 images with 'zerocenter' normalization Classification Output crossentropyex Softmax Fully Connected fully connected layer with 254 output classes ReLU Batch Normalization Convolution 5 filters of size 5x5 convolutions with stride 1 and padding 3 Image Input 8x8x1 or 4x4x1 images with 'zerocenter' normalization
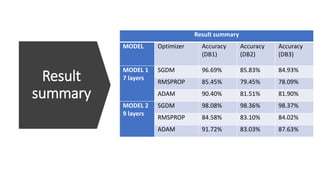
- 84. Result summary Result summary MODEL Optimizer Accuracy (DB1) Accuracy (DB2) Accuracy (DB3) MODEL 1 7 layers SGDM 96.69% 85.83% 84.93% RMSPROP 85.45% 79.45% 78.09% ADAM 90.40% 81.51% 81.90% MODEL 2 9 layers SGDM 98.08% 98.36% 98.37% RMSPROP 84.58% 83.10% 84.02% ADAM 91.72% 83.03% 87.63%
- 85. Best model performanc e evaluation CNN best model performance evaluation Platform Query length 5000 20K 50K 100K 200K GPU Time(s) 6.5 16 39.5 77.8 156 GCUPS 0.0038 0.0248 0.0633 0.1286 0.2563
- 86. Block diagram of testing phase Figure 10. Best model learning progress curve Figure 11. Best model loss curve Sequence 2 Sequence 1 Image Encoding i.e.: reshape input data as DB3 Best Model i.e.: Model 2, DB3, SGDM Labels: 1, 2 …,254 Label Decoding 1 i.e.: '****||||**** ' Result i.e.: 'ACGT||||ACGT ' Decoding 2 i.e.: Replace * by corresponding letter Block diagram of testing phase
- 87. Classical machine learning for global Sequence Alignment 15 ML classifiers for global Sequence Alignment
- 88. Proposed parallel workflow (a) Proposed parallel workflow and (b) traditional NW algorithm sequential workflow
- 89. DESCRIPTION OF DATASETS DESCRIPTION OF DATASETS Dataset1 Dataset2 Dataset3 Dataset4 Dataset5 Dataset 6 Number of Instances (Size) 65536 65536 65536 65536 65483 65483 Input Attributes (Features) 8 (Decimal) 8 (Decimal) 16 (Binary) 16 (Binary) 8 (Decimal) 16 (Binary) Output Classes 254 239 254 239 221 221
- 90. IEEE Dataport
- 93. Data Cleaning Data cleaning involves fixing systematic problems or errors in “messy” data. Using statistics to define normal data and identify outliers. Identifying columns that have the same value or no variance and removing them Identifying duplicate rows of data and removing them. Marking empty values as missing. Imputing missing values using statistics or a learned model.
- 95. FEATURE SELECTION BY USING THE FILTER METHOD (CHI-SQUARED) FOR THE BEST DATASETS FEATURE SELECTION BY USING THE FILTER METHOD (CHI-SQUARED) FOR THE BEST DATASETS χ² (Dataset6) χ² (Dataset3) Feature Number χ² (Dataset6) χ² (Dataset3) Feature Number 497.2075 514.3212 fr3 2335.2803 2928.6962 fr8 478.7286 495.2717 fr11 2318.7507 2912.0966 f16 455.0509 472.0676 fr5 2168.6677 2749.3369 fr4 451.2413 468.8299 f13 2161.9829 2742.2791 fr12 407.9344 430.3850 fr9 2140.9491 2735.2433 fr10 402.7023 425.5596 fr1 2130.2249 2727.2088 fr2 355.1349 374.5339 fr7 2120.4733 2700.1408 fr6 356.3789 375.7883 fr15 2112.1734 2692.3486 fr14
- 96. Feature importance using Xgboost (a) Feature importance for Dataset 5 (b) Feature importance for Dataset 6
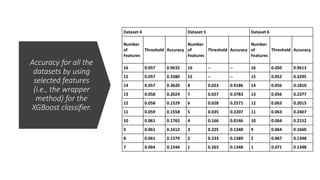
- 97. Accuracy for all the datasets by using selected features (i.e., the wrapper method) for the XGBoost classifier Dataset 1 Dataset 2 Dataset 3 Numb er of Featur es Thresho ld Accura cy Numb er of Featur es Thresho ld Accura cy Numb er of Featur es Thresho ld Accura cy 16 -- -- 16 -- -- 16 0.052 0.9641 15 -- -- 15 -- -- 15 0.056 0.2990 8 0.117 0.9683 8 0.028 0.9564 14 0.057 0.3517 7 0.119 0.3323 7 0.029 0.3830 13 0.057 0.1807 6 0.123 0.2048 6 0.031 0.3097 12 0.059 0.0984 5 0.124 0.1532 5 0.120 0.2158 10 0.061 0.0973 4 0.125 0.0825 4 0.130 0.0258 9 0.063 0.1145 3 0.130 0.0606 3 0.141 0.1131 8 0.065 0.0737 2 0.130 0.0294 2 0.240 0.0322 2 0.069 0.0232 1 0.132 0.0220 1 0.282 0.1332 1 0.070 0.0216
- 98. Accuracy for all the datasets by using selected features (i.e., the wrapper method) for the XGBoost classifier. Dataset 4 Dataset 5 Dataset 6 Number of Features Threshold Accuracy Number of Features Threshold Accuracy Number of Features Threshold Accuracy 16 0.057 0.9632 16 -- -- 16 0.050 0.9613 15 0.057 0.3380 15 -- -- 15 0.052 0.3295 14 0.057 0.3620 8 0.023 0.9186 14 0.056 0.1810 13 0.058 0.2024 7 0.027 0.3783 13 0.056 0.2377 12 0.058 0.1529 6 0.028 0.2571 12 0.063 0.2015 11 0.059 0.1558 5 0.035 0.2207 11 0.063 0.2407 10 0.061 0.1765 4 0.166 0.0146 10 0.064 0.2152 9 0.061 0.1612 3 0.225 0.1348 9 0.064 0.1660 8 0.061 0.1579 2 0.233 0.1389 2 0.067 0.1348 7 0.064 0.1544 1 0.263 0.1348 1 0.071 0.1348
- 99. MODEL HYPER-PARAMETERS Model Model Hyper-parameters 1-Logistic Regression (LR)-LASSO Regularization type: lasso (L1), strength C=1 2-Logistic Regression (LR)-Ridge Regularization type: Ridge (L2), strength C=1 3-Neural Network (MLP) Number of hidden neurons =100, activation=ReLu, solver=SGD, regularization, α=0.0001, number of iterations=1000, replicable training 4-SVM-RBF SVM TYPE=SVM, Cost(C)=1, Regression loss epsilon (Ꜫ)=0.1, Kernel=RBF, g: auto, numerical tolerance=0.001, iteration limit=100 5-SVM –Linear SVM TYPE=SVM, Cost(C)=1, Regression loss epsilon (Ꜫ)=0.1, Kernel=linear, g: auto, numerical tolerance=0.001, iteration limit=100 6-KNeighbors (kNN) Number of neighbors =5, metric=Euclidean, weight=uniform 7-Random Forest Number of trees=10, do not split subsets smaller than= 5 8-Gaussian Naive Bayes (GNB) Default 9-Stochastic Gradient Descent (SGD) Classification loss function =Hinge, regularization method=Ridge(L2), learning rate=constant, initial learning rate=0.01, number of iterations=1000, Tolerance (stopping criterion) =0.001 10-Decision Tree Induce binary tree, min number of instances in leaves =2, do not split subsets smaller than= 5, limit the maximal tree depth=100, stop when majority reaches=95% 11-AdaBoost Base estimator=tree, number of estimators=50, learning rate=1, classification algorithm=SAMME, regression loss function=exponential 12-CN2 rule inducer Rule ordering=ordered, covering algorithm=exclusive, rule search: evaluation measure=Entropy, Beam width=5, Rule Filtering, Minimum rule coverage=1, maximum rule length=5 13-Extra Gradient Boosting (XGBoost) Default 14-Extra Trees Max-depth=4, criterion='entropy', random-state=123 15-Bagging Classifier Base classifier=MLP Classifier, max-samples=0.5, max-features=0.5
- 100. Training accuracy summary results on all the datasets Model Training Time (s) Dataset 1 Accuracy Dataset 2 Accuracy Dataset 3 Accuracy Dataset 4 Accuracy Dataset 5 Accuracy Dataset 6 Accuracy 1-Neural Network (MLP) 24560.971 0.9650 0.9570 0.9847 0.9818 0.9584 0.9838 2-XGBoost 70999 0.9758 0.9168 0.9641 0.9694 0.8929 0.9689 3-AdaBoost 12.294 0.5217 0.5214 0.2912 0.3788 0.5231 0.3802 4-Random Forest 24.251 0.4362 0.4681 0.3449 0.3927 0.4649 0.3877 5-Decision Tree 447.251 0.4025 0.4000 0.2188 0.2493 0.4014 0.2480 6-SVM–RBF 12967.743 0.3486 0.3667 0.8272 0.8415 0.3670 0.8414 7-k-NN 78.96 0.2247 0.2770 0.2565 0.3070 0.2745 0.3089 8-CN2 Rule Inducer 150303.332 0.1946 0.2405 0.1699 0.2110 0.2362 0.2108 9-Extra Trees 67.152 0.0678 0.1391 0.0713 0.1391 0.1392 0.1392 10-GNB 0.573 0.0218 0.0878 0.0090 0.1066 0.1460 0.1403 11-SGD 139.872 0.0147 0.0393 0.0225 0.0484 0.0279 0.0453 12-SVM-Linear 4718.23 0.0095 0.0131 0.0232 0.0241 0.0130 0.0247 13-LR-Ridge 36780.96 0.0069 0.1391 0.0224 0.1398 0.1392 0.1399 14-LR-Lasso 1300.71 0.0068 0.1391 0.0226 0.1395 0.1392 0.1396 15- Bagging Classifier 224.31 0.2481 0.1511 0.1947 0.1402 0.1518 0.1405
- 101. Model Optimizer Classification Accuracy F-measure Precision Recall Neural Network (MLP) ADAM 0.9925 0.9924 0.9924 0.9925 Neural Network (MLP) SGD 0.9847 0.9842 0.9841 0.9847 Neural Network (MLP) L-BFGS-B 0.9842 0.9840 0.9840 0.9842 Model Optimizer Classification Accuracy F-measure Precision Recall Neural Network (MLP) ADAM 0.9927 0.9927 0.9927 0.9927 Neural Network (MLP) SGD 0.9838 0.9833 0.9833 0.9838 Neural Network (MLP) L-BFGS-B 0.9853 0.9853 0.9854 0.9853 Summary of the experiment results of a neural network (i.e., MLP classifier with different optimizers) on Dataset 3 and Dataset 6
- 102. Evaluation result summary of a neural network (i.e., MLP classifier (for Datasets 3T and 6T (testing phase) Model Optimizer Classification Accuracy F-measure Precision Recall Neural Network (MLP)- Dataset 3T ADAM 0.859 0.859 0.859 0.859 Neural Network (MLP)- Dataset 6T ADAM 0.860 0.859 0.859 0.860
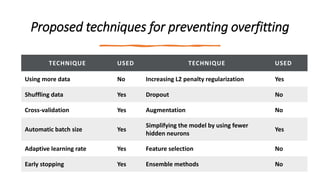
- 103. Proposed techniques for preventing overfitting TECHNIQUE USED TECHNIQUE USED Using more data No Increasing L2 penalty regularization Yes Shuffling data Yes Dropout No Cross-validation Yes Augmentation No Automatic batch size Yes Simplifying the model by using fewer hidden neurons Yes Adaptive learning rate Yes Feature selection No Early stopping Yes Ensemble methods No
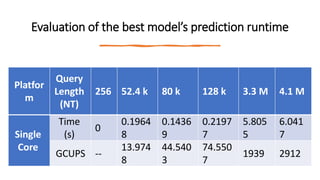
- 104. Evaluation of the best model’s prediction runtime Platfor m Query Length (NT) 256 52.4 k 80 k 128 k 3.3 M 4.1 M Single Core Time (s) 0 0.1964 8 0.1436 9 0.2197 7 5.805 5 6.041 7 GCUPS -- 13.974 8 44.540 3 74.550 7 1939 2912
- 105. Performance comparison with other state-of-the-art implementations REFERENCE YEAR SEQUENCE PAIRS TIME (S) GCUPS [16] 2013 1024 NTs 7.065 1.4842×10−4 [38] 2014 D4.4 vs. D4.6 203 100.7 [18] 2015 14336 NTs 12 0.0171 [22] 2019 20 k NTs 0.5995 0.6672 [23] 2019 50 k NTs 1157.8305 (19 min) 0.0022 [20] 2020 3 M NTs 29499 (492 min) 0.3051 Proposed 52.4 k NTs (Dataset 3T) 0.19648 13.9748 Proposed 4.1 M NTs (Dataset 4.1 M) 6.0417 2912
- 106. Strategy for testing new data
- 108. Conclusion • Most of the previous studies aimed to accelerate the alignment algorithms in different ways without providing any effective solution for sequential process problems. Our proposed algorithms depend on the parallelization of common alignment algorithms for DNA sequences under certain limitations to overcome the main problems of DP and hardware implementation. It can also be applied to RNA. This technique can be applied to any other local or global alignment method and for short as well as very long sequences. The proposed technique using MATLAB achieves considerably better elapsed time and GCUPS than the state-of-the-art technique for local and global alignment. FPGA is demonstrated as a cost-effective, energy-efficient platform for the implementation of sequence alignment algorithms. This study presents a 65,536 × 48 ROM (LUT)-based hardware implementation for the local alignment algorithm using VHDL language. • Our proposed implementation requires only one clock cycle to obtain full alignment working on a 400 MHZ frequency. The same technique (ROM-based) design can also be used for the NW algorithm, but we opt to use another approach and check its performance. • The combinational circuit design for SW algorithm achieves maximum path delays of 15 and 7.511 ns (as in the fourth design) for NW algorithm on XilinxVirtex6 FPGA. Moreover, the estimated power is approximately the same for the two alignment algorithms. The improvement that occurs in NW alignment (maximum path delay in combinational circuit design) over SW alignment algorithm is due to our minimization procedure (reduction techniques used in NW alignment). The third design (NW algorithm) achieves the least number of logic circuits used (21,592) based on two reduction techniques and logic minimization techniques. A customized CNN model is used for the software implementation of a NW algorithm, and 98.3% accuracy is achieved. This accuracy can reach 99.21% in absence of a dropout layer.
- 109. Future work • Using different opening gap values in NW design does not substantially affect the HW performance as well as the number of characters representing the alignment array (still 18 characters), but reviewing and standardizing the alignment array can (i.e., use a single pattern for all full-mismatch conditions [’****::::****’] or a single symbol to represent mismatch condition [colons only] instead of using two symbols [space and colon]. In addition, this standardization will reduce the number of classes (the target for the CNN model) and enhance the performance of the CNN model. ML techniques used in this study do not a achieve reasonable accuracy which can be improved later by using python auto-ML libraries like Auto-Sklearn, TPOT, HyperOpt, and AutoKeras. Auto-ML libraries can also improve CNN model accuracy by tuning the hyperparameters with the use of recent methods such as Bayesian optimization, or automating feature selection, preprocessing, and construction or searching for the best architecture. Our CNN model can be later implemented using FPGA, which may improve hardware design speed, device usage utility, and performance.
Editor's Notes
- #3: https://www.sciencedirect.com/science/article/abs/pii/S0045790621001178
- #14: Deoxyribonucleic acid (DNA) and Ribonucleic acid (RNA) are nucleic acids consist of a nucleo-base, a pentose sugar and a phosphate group.
- #36: Maria Kim ,‘Accelerating Next Generation Genome Reassembly in FPGAs: Alignment Using Dynamic Programming Algorithms’ ,master thesis , University of Washington ,2011.
- #43: In addition, the type of alignment algorithm (SW or NW) affects the alignment array. For example, to align (AACC, CCAA) using NW algorithm the alignment array will have 18 characters as follows ('AACC-- || --CCAA') for our proposed value of opening gap or 12 characters ('AACC CCAA') when opening gap equals 8 (default S/W value). As another example to align (AAAC, AACA) using NW algorithm, the alignment array will consist of 18 characters ('AACC-- || --CCAA') for our proposed value of opening gap, 12 characters ('AAAC|| AACA') when opening gap equals 8, and 9 characters ('AAC|||AAC') when using the SW algorithm. Note that, space as colon between two sequences (same meaning) is used to represent mismatches.
- #46: BLOSUM series (Henikoff S. & Henikoff JG., PNAS, 1992) Blocks Substitution Matrix. A substitution matrix in which scores for each position are derived from observations of the frequencies of substitutions in blocks of local alignments in related proteins. Each matrix is tailored to a particular evolutionary distance. In the BLOSUM62 matrix, for example, the alignment from which scores were derived was created using sequences sharing no more than 62% identity. Sequences more identical than 62% are represented by a single sequence in the alignment so as to avoid over-weighting closely related family members. !Based on alignments in the BLOCKS database !Standard matrix: BLOSUM62
- #91: https://ieee-dataport.org/documents/dna-sequence-alignment-datasets-based-nw-algorithm.
- #95: Feature Selection: Select a subset of input features from the dataset. Unsupervised: Do not use the target variable (e.g. remove redundant variables). Correlation Supervised: Use the target variable (e.g. remove irrelevant variables). Wrapper: Search for well-performing subsets of features. RFE Filter: Select subsets of features based on their relationship with the target. Statistical Methods (chi-squared, relief, F-statistic, mRMR, and information gain) Feature Importance Methods Intrinsic: Algorithms that perform automatic feature selection during training. Decision Trees Dimensionality Reduction: Project input data into a lower-dimensional feature space.






![Problem Definition
(1) The number of sequences is large, and each of their
lengths can be very long.
(2) Table II shows that the algorithms used to align the
sequences requires O(MN) calculation steps and
consumes O(MN) time (M and N are the lengths of the
two input sequences).
(3) Basic sequence alignment algorithms are internally
dependent on the sequential process.
(4) Hardware implementation of sequence alignment
algorithms does not present an effective solution for
sequential process problems, which affects system
speed.
(5) Practical problems exist owing to parallel
implementations, such as the communication overheads.
(6) Dynamic algorithms guarantee optimal alignment,
but they are slower than FASTA and BLAST, and require
extensive computation time and memory because of the
sequential processes. Although FASTA and BLAST are
fast, they do not guarantee optimum alignment. Our
proposed algorithms
TABLE II COMPARE SPACE AND TIME COMPLEXITY FOR DP
ALGORITHMS [1]
Algorithm Type Space
Complexity
Time
Complexity
SW Local-linear
gap penalty
O(MN) O(MN)
Gotoh Local-affine
gap penalty
O(MN) O(MN)
Miller–
Myers
Local-affine
gap penalty
O(M+N) O(NM)
NW Global-linear
gap penalty
O(MN) O(MN)
Hirschberg Global-linear
gap penalty
O(M+N) O(MN)](https://image.slidesharecdn.com/presentation11-7-2021-210816162951/85/Implementation-of-DNA-sequence-alignment-algorithms-using-Fpga-ML-and-CNN-6-320.jpg)






































![Performance comparison with other state-of-the-art implementations
Performance comparison with other state-of-the-art implementations
Paper Year Platform Sequence Pairs Time (s) GCUPS
[15] 2014 1 Xeon Phi D4.4 vs D4.6M 700 29.2
[15] 2014 2 Xeon Phis D4.4 vs D4.6M 396 51.7
[15] 2014 4 Xeon Phis D4.4 vs D4.6M 203 100.7
[16] 2014 Intel® Core™ i7-3770 CPU @ 3.40GHz×8. 256NT vs 265NT 0.317 --
[17] 2019 2×Xeon Gold 6138 Max Query length=5478 -- 734
Ours (SW
algorithm)
Intel Core I7-9750H 6-cores 2.60 GHz
CPUs
Same sequence pairs in [21], after
cropping the second sequence to 4.4M
7.8607 2462.9
Ours (SW
algorithm)
Intel Core I7-9750H 6-cores 2.60 GHz
CPUs
256NT vs 265NT same length as in [15] 0.1745 --
Ours (NW
algorithm)
Intel Core I7-9750H 6-cores 2.60 GHz
CPUs
Same sequence pairs in [21], after
cropping the second sequence to 4.4M
8.3676 2313.7](https://image.slidesharecdn.com/presentation11-7-2021-210816162951/85/Implementation-of-DNA-sequence-alignment-algorithms-using-Fpga-ML-and-CNN-51-320.jpg)









![COMPARISON BETWEEN WIDELY USED LOGIC FUNCTION MINIMIZATION
METHODS
COMPARISON BETWEEN WIDELY USED LOGIC FUNCTION MINIMIZATION METHODS
Karnaugh Map (K-Map) Quine–McCluskey (QM) Espresso algorithm
Definition
Method for simplifying Boolean
algebra expressions
Known as the tabulation method or the technique of prime
implicants, it is functionally similar to Karnaugh Maps.
A radically different approach,
the algorithm manipulates "cubes," performing the product
terms in the ON-, DC-, and OFF covers iteratively.
Features
Four variables.
Unsuitable for more than 6 input variables
Tedious and error-prone process
It can be performed manually and does
not support more than 8 input bits.
Challenge to be implemented in computer
programs[66][67].
K-Map is not suitable for our algorithms
because these have 16 input bits.
Can still be performed manually on paper
Scales to many variables (Can handle up to 40 variables)
One of the highly effective techniques for simplifying Boolean
expressions.
More convenient to be implemented in computer programs.
Has a tabular form that makes it more efficient for use in
computer algorithms
Has a settled methodology to test whether the minimal form of a
Boolean function has been attained.
For a larger number of input variables, QM is more effective in
minimizing logic functions than K-Map[66][67].
Is not guaranteed to be the global minimum.
Practically, it is very closely approximated and free from
redundancy.
Computationally efficient in terms of both memory
requirement and time than the other methods (K-Map and
QM) by several orders of magnitude
Used as a standard logic expression minimization in logic
synthesis tools](https://image.slidesharecdn.com/presentation11-7-2021-210816162951/85/Implementation-of-DNA-sequence-alignment-algorithms-using-Fpga-ML-and-CNN-62-320.jpg)










![Performance evaluation(1)
COMPARISON OF THE PERFORMANCE OF VARIED SINGLE-DEVICE IMPLEMENTATIONS OF THE SW/NW ALGORITHM
Paper Year Algorithm Circuit Type Technique Device Frequency
(MHz)
Time
(ns)
GCUPS
[21] 2009 SW/NW Sequential Systolic cell Xilinx Virtex-4 FX100 100 -- 25.6
[22] 2014 SW Sequential Systolic cell Altera Stratix IV
EP4SGX230
57.9 17.27 3.71
[23] 2016 SW Sequential Systolic cell Xilinx XC3S1600E 98.7 10.13 23.79
[24] 2017 SW Sequential Systolic cell -- 250 -- --
[24] 2017 SW Sequential Systolic cell -- 250 -- --
Ours -- SW -Design1 Sequential LUT Xilinx Virtex 6
XC6VLX240T
400 2.499 25.6102
Ours -- NW -Design 4 Combinational LUT Xilinx Virtex 6
XC6VLX240T
133 7.511 8.5333](https://image.slidesharecdn.com/presentation11-7-2021-210816162951/85/Implementation-of-DNA-sequence-alignment-algorithms-using-Fpga-ML-and-CNN-74-320.jpg)
![Performance evaluation(2)
COMPARISON OF USAGE OF FPGA RESOURCES
Paper Year Device Frequency
(MHz)
LUT BRAM Others
[25] 2017 -- 250 3056/ 537600 (0.57%) 94/3456 (2.72%) FF- 2468 (0.23%)
[26] 2018 Stratix IV
EP4SGX230KF40C2
125 70,839 / 182,400
(39%)
1,834,562 /
14,625,792 (13%)
Total Registers 73,850
[27] 2018 Kintex7 XC7K480T
FFG901-3
100 17462 (8%) -- FF-1402 (1%)
Latches-1483 (1%)
LUT-FF pairs 216 (5%)
Ours -- Xilinx Virtex 6
XC6VLX240T
400 0% 96/416 (23%) 0%
Ours -- Xilinx Virtex 6
XC6VLX240T
-- 21872 /150720 (14%) 0% 0%](https://image.slidesharecdn.com/presentation11-7-2021-210816162951/85/Implementation-of-DNA-sequence-alignment-algorithms-using-Fpga-ML-and-CNN-75-320.jpg)
![Performance evaluation(3)
COMPARISON OF NUMBER OF OPERATIONS, STEPS, OR CLOCK CYCLES FOR COMPLETE ALIGNMENT
Paper Year Algorithm Technique # of Operations for complete
alignment
[9] 2012 SW Systolic array (N+M−1+Trace-back operations)
[28] 2018 SW Systolic array (N+M−1+Trace-back operations)
[29] 2020 SW Systolic array (N+M−1+Trace-back operations)
Ours -- SW/NW LUT (N/4)](https://image.slidesharecdn.com/presentation11-7-2021-210816162951/85/Implementation-of-DNA-sequence-alignment-algorithms-using-Fpga-ML-and-CNN-76-320.jpg)





















![Performance comparison with other state-of-the-art
implementations
REFERENCE YEAR SEQUENCE PAIRS TIME (S) GCUPS
[16] 2013 1024 NTs 7.065 1.4842×10−4
[38] 2014 D4.4 vs. D4.6 203 100.7
[18] 2015 14336 NTs 12 0.0171
[22] 2019 20 k NTs 0.5995 0.6672
[23] 2019 50 k NTs 1157.8305 (19 min) 0.0022
[20] 2020 3 M NTs 29499 (492 min) 0.3051
Proposed 52.4 k NTs (Dataset 3T) 0.19648 13.9748
Proposed 4.1 M NTs (Dataset 4.1 M) 6.0417 2912](https://image.slidesharecdn.com/presentation11-7-2021-210816162951/85/Implementation-of-DNA-sequence-alignment-algorithms-using-Fpga-ML-and-CNN-105-320.jpg)



![Future work
• Using different opening gap values in NW design does not
substantially affect the HW performance as well as the number of
characters representing the alignment array (still 18 characters), but
reviewing and standardizing the alignment array can (i.e., use a
single pattern for all full-mismatch conditions [’****::::****’] or a single
symbol to represent mismatch condition [colons only] instead of
using two symbols [space and colon]. In addition, this
standardization will reduce the number of classes (the target for the
CNN model) and enhance the performance of the CNN model. ML
techniques used in this study do not a achieve reasonable accuracy
which can be improved later by using python auto-ML libraries like
Auto-Sklearn, TPOT, HyperOpt, and AutoKeras. Auto-ML libraries
can also improve CNN model accuracy by tuning the
hyperparameters with the use of recent methods such as Bayesian
optimization, or automating feature selection, preprocessing, and
construction or searching for the best architecture. Our CNN model
can be later implemented using FPGA, which may improve
hardware design speed, device usage utility, and performance.](https://image.slidesharecdn.com/presentation11-7-2021-210816162951/85/Implementation-of-DNA-sequence-alignment-algorithms-using-Fpga-ML-and-CNN-109-320.jpg)