Protein and RNA alignment and analysis with Jalview 2.8.2 and JABA 2.1
- 1. Protein and RNA alignment and analysis with Jalview 2.8.2 and JABA 2.1 TT18 and Poster B38 Jim Procter [email protected] Jalview Coordinator, Barton Group College of Life Sciences, University of Dundee, UK.
- 2. Standalone or web based Java alignment viewer and editor & annotation, tree, and structure Available at www.jalview.org
- 3. One alignment, many views Sequence features highlight key regions like functional sites Alignment annotation area shows graphs and symbols from calculations and manual curation
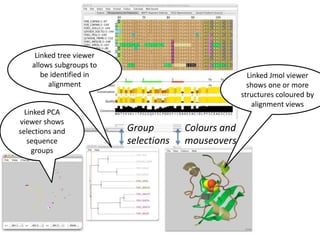
- 4. Linked tree viewer allows subgroups to be identified in alignment Group selections Colours and mouseovers Linked Jmol viewer shows one or more structures coloured by alignment views Linked PCA viewer shows selections and sequence groups
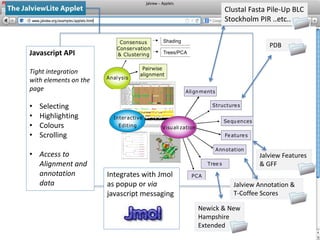
- 5. Interactive Editing Visuali zation Alignments Structures Features Annotation Tree s Sequences PCA Analysis Pairwise alignment Consensus Conservation & Clustering Shading Trees/PCA Newick & New Hampshire Extended Jalview Annotation & T-Coffee Scores Jalview Features & GFF Clustal Fasta Pile-Up BLC Stockholm PIR ..etc.. PDB Javascript API Tight integration with elements on the page • Selecting • Highlighting • Colours • Scrolling • Access to Alignment and annotation data Integrates with Jmol as popup or via javascript messaging
- 6. JavascriptAPI Jalview comes in two flavours
- 8. VARNA Visual Analysis of RNA http://jmol.sourceforge.net/ http://varna.lri.fr/ Desktop Structure Visualization 3D structures and 2D RNA diagrams
- 9. Clustal Fasta Pile-Up BLC Stockholm PIR ..etc.. Newick & New Hampshire Extended Jalview Annotation & T-Coffee Scores Jalview Features & GFF PDB Distributed Annotation System Interactive Editing Visuali zation Alignments Structures Features Annotation Tree s Sequences PCA Figure Generation Clickable HTML Clic kable HTML Line ArtLine Art ImagesImages Analysis Pairwise alignment Consensus Conservation & Clustering Shading Trees/PCA
- 10. JAVA BIOINFORMATICS ANALYSIS WEB SERVICES Public services and downloads at www.compbio.dundee.ac.uk/jabaws /
- 11. The JABAWS Java Client Library Jalview Web Service GUI JABAWS command line interface JABAWS Java Client ClustalW Mafft Multiple alignmentProtein Disorder Alignment Analysis JABA Web Server
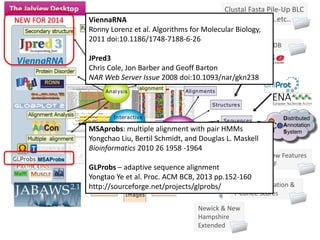
- 13. Clustal Fasta Pile-Up BLC Stockholm PIR ..etc.. Newick & New Hampshire Extended Jalview Annotation & T-Coffee Scores Jalview Features & GFF PDB Distributed Annotation System Interactive Editing Visuali zation Alignments Structures Features Annotation Tree s Sequences PCA Figure Generation Clickable HTML Clic kable HTML Line ArtLine Art ImagesImages Analysis Pairwise alignment Consensus Conservation & Clustering Shading Trees/PCA NEW FOR 2014 MSAprobs: multiple alignment with pair HMMs Yongchao Liu, Bertil Schmidt, and Douglas L. Maskell Bioinformatics 2010 26 1958 -1964 GLProbs – adaptive sequence alignment Yongtao Ye et al. Proc. ACM BCB, 2013 pp.152-160 http://sourceforge.net/projects/glprobs/ ViennaRNA Ronny Lorenz et al. Algorithms for Molecular Biology, 2011 doi:10.1186/1748-7188-6-26 JPred3 Chris Cole, Jon Barber and Geoff Barton NAR Web Server Issue 2008 doi:10.1093/nar/gkn238
- 14. Native JABAWSinstalls on arange of platforms powered by JABAWSAmazon Machine Image on EC2 Includes all 3rd party source & binaries and databases Reconfigurable parameters, presets & execution limits Web UI for installation and status checks
- 15. Released along-side Jalview 2.8.1 on 5th June 2014 New optional downloads: 2.3G Jpred database Provided as tarball/virtual disk for native and VM installs http://www.compbio.dundee.ac.uk/jabaws/
- 16. THE JALVIEW 2.8 SERIES
- 17. IT’S (NEARLY) ALL ABOUT STRUCTURE!
- 18. Jalview 2.8 and RNA 2nd-ary Structure Structure Consensus Logo: Shows base pair distribution at each paired position in a given RNA secondary structure. Linked VARNA RNA Secondary Structure viewer and editor. RALEE style colouring highlights distinct stems and helices
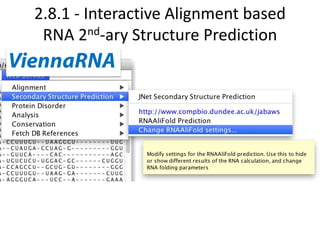
- 19. 2.8.1 - Interactive Alignment based RNA 2nd-ary Structure Prediction
- 20. 2.8.1 - Interactive Alignment based RNA 2nd-ary Structure Prediction • Shares framework with AACon consensus client • Predictions update when alignment changes • settings & results saved in Jalview project Implemented by our 2013 Summer student
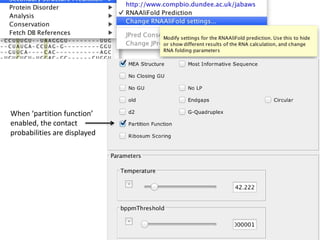
- 21. When ‘partition function’ enabled, the contact probabilities are displayed
- 22. Tooltips provide additional RNAAliFold information
- 23. Protein Secondary Structure Prediction • Neural network trained on amino acid profiles – Predicts Helix, shEet, or Coil based on sliding window • Also predicts coiled coils and surface accessibilities • Server can take – Single Sequence • Service computes profile with PSI-Blast – Alignment • Service uses MSA to calculate profile for prediction SLOW FAST! Interactive JPred3 MSA annotation now in 2.8.2 alpha
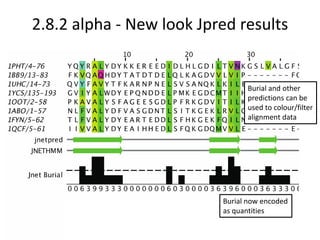
- 24. 2.8.2 alpha - New look Jpred results Burial now encoded as quantities Burial and other predictions can be used to colour/filter alignment data
- 25. Secondary structure from 3D data • Jmol includes a Java port of DSSP – Courtesy of the Vriend Lab • Jalview 2.8.2 now employs Jmol to parse PDB data – Display residue level structure data on sequences http://jmol.sourceforge.net/
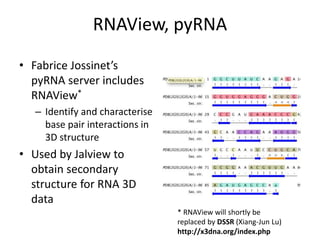
- 26. RNAView, pyRNA • Fabrice Jossinet’s pyRNA server includes RNAView* – Identify and characterise base pair interactions in 3D structure • Used by Jalview to obtain secondary structure for RNA 3D data * RNAView will shortly be replaced by DSSR (Xiang-Jun Lu) http://x3dna.org/index.php
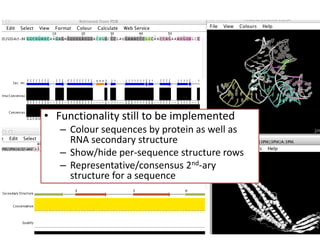
- 27. • Functionality still to be implemented – Colour sequences by protein as well as RNA secondary structure – Show/hide per-sequence structure rows – Representative/consensus 2nd-ary structure for a sequence
- 28. Things I haven’t talked about … Currently available in v 2.8.1 • Internationalisation (Spanish, so far) • View flanking regions (Proteomics) • More score models for PCA/Trees • View ‘representative structures’ • Select columns by feature..
- 29. Select column by feature
- 30. Roadmap for Jalview 2.8.2 E.T.A September 2014 • Interactive consensus secondary structure annotation for protein alignments – JABA Jpred MSA prediction mode • Structure based RNA + Protein annotation – Jmol DSSP and XRNA 2.5D • Prototype Chimera/Jalview interconnect • Feature/Annotation based trees/PCA – Cluster by • Secondary structure • Displayed sequence annotation
- 31. Michele Clamp Harvard & MIT USA. James Cuff Harvard & MIT USA. The Jalview & JABA Saga Steve Searle Sanger, UK Andrew Waterhouse U. Basel. Jalview Version 2 2005 Jalview Version 1 1997 Jim Procter (still in Dundee!) David Martin CLSLT2004 Jalview 1 published. 2009 Jalview 2 published 2.1 2.2 2.3 2.4 2.5 2.6.1 2.7 Peter Troshin NHS VAMSAS Sasha Sherstnev GSK 2.8.1 Jalview BBR 2009-2014 JABA BBR ‘13-’18 JABA 1 paper 2011
- 32. Lines of Code x10-3 0 200 250 300 2009 2010 2011 2012 2.4.0b2 2.5.1 2.6.1 2.6 2.7 2.8 2.4.0b2 2.5 Development highlights since 2009 ClustalW Mafft New Contributions from Lauren Lui & Jan Engelhart (Google Summer of Code) Anne Menard, Yann Ponty (Paris Sud funding) Paolo Di Tomasso & Natasha Sherstnev David Roldán Martinez Thousands of Lines of Code
- 33. The next 5 Years
- 34. And the Saga continues… Suzanne Duce Training & Outreach Jalview & JABA Jalview Visual Analytics Scientist/Developers Alexey Drozdetskiy Scientist/Developer JABA