KNIME tutorial
- 1. Day 4: KNIME Tutorial George Papadatos, PhD Francis Atkinson, PhD ChEMBL group
- 2. Outline • Introduc>on to KNIME • Basic components • Desktop, nodes, dialogs, workflows • Exercise • Compound selec>on for focused screening • Read chemical data • Calculate proper>es • Apply drug-‐ and lead-‐ likeness filters • Remove “nasty” compounds • Pick diverse molecules • Visualize results and plot proper>es 2 05/07/2012 Resources for Computa5onal Drug Design
- 3. What is KNIME? • KNIME = Konstanz Informa>on Miner • Developed at University of Konstanz in Germany • Desktop version available free of charge (Open Source) • Modular plaWorm for building and execu>ng workflows using predefined components, called nodes • Core func>onality available for tasks such as standard data mining, analysis and manipula>on • Extra features and func>onality available in KNIME through extensions from various groups and vendors • WriYen in Java based on the Eclipse SDK plaWorm 3 05/07/2012 Resources for Computa5onal Drug Design
- 4. KNIME resources • Web pages (documenta>on) • www.knime.org | tech.knime.org | tech.knime.org/installa>on-‐0 • Downloads • knime.org/download-‐desktop • Community forum • tech.knime.org/forum • KNIME User Training Manual • Books and white papers • knime.org/node/33079 • Myself • [email protected] 4 05/07/2012 Resources for Computa5onal Drug Design
- 5. What can you do with KNIME? • Data manipula>on and analysis • File & database I/O, sor>ng, filtering, grouping, joining, pivo>ng • Data mining / machine learning • R, WEKA, interac>ve plofng • Chemoinforma>cs • Conversions, similarity, clustering, (Q)SAR analysis, reac>on enumera>on • Scrip>ng integra>on • R, Perl, Python, Matlab, Octave, Groovy • Repor>ng • Much more • Bioinforma>cs, image analysis, network & text mining 5 05/07/2012 Resources for Computa5onal Drug Design
- 6. Community contributions • hYp://tech.knime.org/community • Chemoinforma>cs • CDK (EBI), RDKit (Novar>s), Indigo (GGA), ErlWood (Eli Lilly), Enalos (NovaMechanics) • Bioinforma>cs • HCS (MPI), NGS (Konstanz) • Text mining • Palladian • Integra>on • Python, Perl, R, Groovy, Matlab (MPI), PDB web services client (Vernalis) 6 05/07/2012 Resources for Computa5onal Drug Design
- 7. Installation & updates • Download and unzip KNIME • No further setup required • Addi>onal nodes aker first launch • knime.ini contains arguments & parameters for launch • New sokware (nodes) from update sites • hYp://tech.knime.org/update/community-‐contribu>ons/release • Workflows and data are stored in a workspace • /Users/georgep/knime/workspace_mac_new • C:knime_2.5.4workspace • Customiza>on in: FileàPreferencesàKNIME 7 05/07/2012 Resources for Computa5onal Drug Design
- 8. Auto-‐layout Execute Execute all nodes KNIME Workbench Node descrip>on tabs workflow projects favorite nodes public server workflow editor node repository outline console 8 05/07/2012 Resources for Computa5onal Drug Design
- 9. KNIME nodes: Overview Node = basic processing unit of KNIME workflow which performs a par>cular task Input port(s) – on the lek of icon Title Output port(s) – on the right of icon Icon Status display (‘traffic lights’) Right-‐click menu Sequence number • Red (not ready) To configure and • Amber (ready) execute the node, • Green (executed) display the output views, edit the • Blue bar during execu>on node, and display (with percentage or flashing) data for the ports 9 05/07/2012 Resources for Computa5onal Drug Design
- 10. KNIME nodes: Dialogs Double click to configure… Configura>on menus for selected nodes Explicit column type 10 05/07/2012 Resources for Computa5onal Drug Design
- 11. An example completed workGlow • Workflows can be imported and exported as .zip files • With or without the underlying data • File à Import KNIME workflow… • File à Export KNIME workflow… 11 05/07/2012 Resources for Computa5onal Drug Design
- 12. Any questions so far? 12 05/07/2012 Resources for Computa5onal Drug Design
- 13. Compound selection for focused screening 1. Read chemical data 2. Remove duplicates • Iden>ty ensured by InChi keys 3. Filter out compounds in ChEMBL • Iden>ty ensured by InChI keys 4. Calculate phys/chem proper>es 5. Apply drug-‐ and lead-‐likeness filters 6. Apply more filters (e.g. remove solubility liabili>es) 7. Apply substructural filters (PAINS subset) 8. Pick diverse molecules 13 05/07/2012 Resources for Computa5onal Drug Design
- 14. Your objective 14 05/07/2012 Resources for Computa5onal Drug Design
- 15. First steps -‐ I • Locate the directory with today’s material 1 2 • Copy and paste it to your desktop • You can take it with you too • Open the presenta>on file • Import the FocusedScreeningSelec>on.zip to KNIME • Menu à File à Import workflow to KNIME 3 15 05/07/2012 Resources for Computa5onal Drug Design
- 16. First steps -‐ II • Open a new workflow • Right click on the workflow projects area 1 2 3 16 05/07/2012 Resources for Computa5onal Drug Design
- 17. Part 1: Reading and cleaning up 17 05/07/2012 Resources for Computa5onal Drug Design
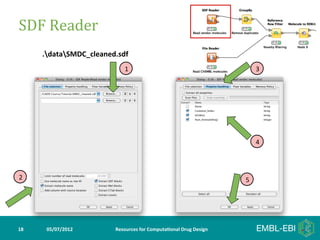
- 18. SDF Reader .dataSMDC_cleaned.sdf 1 3 4 2 5 18 05/07/2012 Resources for Computa5onal Drug Design
- 19. Inspect the structures… Right click on the node 19 05/07/2012 Resources for Computa5onal Drug Design
- 20. GroupBy 1 3 2 5 4 20 05/07/2012 Resources for Computa5onal Drug Design
- 21. GroupBy Example Name Course Grade George German 68 George Maths 86 George Physics 99 Group by Name and Group by Name and then take first row then average Grade Name Course (first) Grade (first) Name Grade (avg.) George German 68 George 84.33 21 05/07/2012 Resources for Computa5onal Drug Design
- 22. File Reader 1 .dataall_human_chembl.csv 2 3 22 05/07/2012 Resources for Computa5onal Drug Design
- 23. Reference Row Filter 23 05/07/2012 Resources for Computa5onal Drug Design
- 24. Molecule to RDKit 24 05/07/2012 Resources for Computa5onal Drug Design
- 25. Any questions so far? 25 05/07/2012 Resources for Computa5onal Drug Design
- 26. Part 2: Property-‐based Giltering 26 05/07/2012 Resources for Computa5onal Drug Design
- 27. Descriptor Calculation 1 2 3 27 05/07/2012 Resources for Computa5onal Drug Design
- 28. Java Snippet 1 .codeLipinski.txt 3 2 28 05/07/2012 Resources for Computa5onal Drug Design
- 29. Numeric Row Splitter 29 05/07/2012 Resources for Computa5onal Drug Design
- 30. Inspect the Lipinski fails… Right click on the node 30 05/07/2012 Resources for Computa5onal Drug Design
- 31. Java Snippet 1 .codeOprea.txt 3 2 31 05/07/2012 Resources for Computa5onal Drug Design
- 32. Numeric Row Splitter 32 05/07/2012 Resources for Computa5onal Drug Design
- 33. Inspect the Oprea fails… Right click on the node 33 05/07/2012 Resources for Computa5onal Drug Design
- 34. Numeric Row Splitter 34 05/07/2012 Resources for Computa5onal Drug Design
- 35. Inspect the Solubility fails… Right click on the node 35 05/07/2012 Resources for Computa5onal Drug Design
- 36. Any questions so far? 36 05/07/2012 Resources for Computa5onal Drug Design
- 37. Part 3: Substructure-‐based Giltering 37 05/07/2012 Resources for Computa5onal Drug Design
- 38. Molecule to Indigo 38 05/07/2012 Resources for Computa5onal Drug Design
- 39. File reader .dataPAINS_clean_half.sdf 39 05/07/2012 Resources for Computa5onal Drug Design
- 40. Query Molecule to Indigo 40 05/07/2012 Resources for Computa5onal Drug Design
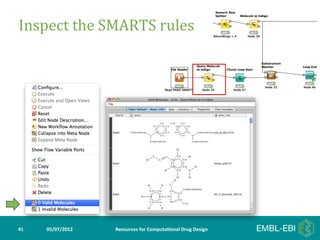
- 41. Inspect the SMARTS rules 41 05/07/2012 Resources for Computa5onal Drug Design
- 42. Chunk Loop Start 42 05/07/2012 Resources for Computa5onal Drug Design
- 43. Substructure Matcher 43 05/07/2012 Resources for Computa5onal Drug Design
- 44. Loop End 44 05/07/2012 Resources for Computa5onal Drug Design
- 45. Inspect matched structures… Right click on the node 45 05/07/2012 Resources for Computa5onal Drug Design
- 46. Reference Row Filter 46 05/07/2012 Resources for Computa5onal Drug Design
- 47. Any questions so far? 47 05/07/2012 Resources for Computa5onal Drug Design
- 48. Part 4: Diversity picking and plotting 48 05/07/2012 Resources for Computa5onal Drug Design
- 49. RDKit Fingerprint 49 05/07/2012 Resources for Computa5onal Drug Design
- 50. Inspect the Gingerprints… Right click on the node 50 05/07/2012 Resources for Computa5onal Drug Design
- 51. RDKit Diversity Picker 51 05/07/2012 Resources for Computa5onal Drug Design
- 52. 2D/3D Scatterplot 52 05/07/2012 Resources for Computa5onal Drug Design
- 53. Inspect the plot… Right click on the node 53 05/07/2012 Resources for Computa5onal Drug Design
- 54. Any questions so far? 54 05/07/2012 Resources for Computa5onal Drug Design
- 55. Conclusions • Compound selec>on for focused screening • Theory and prac>ce • Typical scenario • KNIME • Open and free • Chemoinforma>cs toolkits • Erl Wood, RDKit and Indigo • Not perfect 55 05/07/2012 Resources for Computa5onal Drug Design
- 56. Further reading • Open data and tools 1. A freeJ. J.; Sterling, T.; Mysinger, M. M.; Bolstad, E. S.; Coleman, R. G., ZINC: Irwin, tool to discover chemistry for biology. Journal of Chemical Information and Modeling 2012 ASAP. 2. Saubern, S.; Guha, R.; Baell, J. B., KNIME workflow to assess PAINS filters in SMARTS format. Comparison of RDKit and Indigo cheminformatics libraries. Molecular Informatics 2011, 30, (10), 847-850. 3. Barnes, M. R.; Harland, L.; Foord, S. M.; Hall, M. D.; Dix, I.; Thomas, S.; Williams-Jones, B. I.; Brouwer, C. R., Lowering industry firewalls: pre- competitive informatics initiatives in drug discovery. Nature Reviews Drug Discovery 2009, 8, (9), 701-708. 4. Berthold, M. R.; Cebron, N.; Dill, F.; Gabriel, T. R.; Kötter, T.; Meinl, T.; Ohl, P.; Sieb, C.; Thiel, K.; Wiswedel, B., KNIME: The Konstanz Information Miner. In Data Analysis, Machine Learning and Applications, Preisach, C.; Burkhardt, H.; Schmidt-Thieme, L.; Decker, R., Eds. Springer: Berlin, 2008; pp 319-326. 5. Tiwari, A.; Sekhar, A. K. T., Workflow based framework for life science informatics. Computational Biology and Chemistry 2007, 31, (5-6), 305-319. 56 05/07/2012 Resources for Computa5onal Drug Design
- 57. Further reading • High throughput screening 1. Bajorath, J., Integration of virtual and high-throughput screening. Nature Reviews Drug Discovery 2002, 1, (11), 882-894. 2. Harper, G.; Pickett, S. D.; Green, D. V. S., Design of a compound screening collection for use in High Throughput Screening. Combinatorial Chemistry & High Throughput Screening 2004, 7, (1), 63-70. • Lead-‐ and drug-‐likeness 1. Chuprina, A.; Lukin, O.; Demoiseaux, R.; Buzko, A.; Shivanyuk, A., Drug- and lead-likeness, target class, and molecular diversity analysis of 7.9 million commercially available organic compounds provided by 29 suppliers. Journal of Chemical Information and Modeling 2010, 50, (4), 470-479. 2. Lipinski, C. A., Lead- and drug-like compounds: the rule-of-five revolution. Drug Discovery Today: Technologies 2004, 1, (4), 337-341. 3. Oprea, T. I.; Davis, A. M.; Teague, S. J.; Leeson, P. D., Is there a difference between leads and drugs? A historical perspective. Journal of Chemical Information and Computer Sciences 2001, 41, (5), 1308-1315. 57 05/07/2012 Resources for Computa5onal Drug Design
- 58. Further reading • Physicochemical proper>es and drug discovery 1. Brüstle, M.; Beck, B.; Schindler, T.; King, W.; Mitchell, T.; Clark, T., Descriptors, physical properties, and drug-likeness. Journal of Medicinal Chemistry 2002, 45, (16), 3345-3355. 2. Hill, A. P.; Young, R. J., Getting physical in drug discovery: A contemporary perspective on solubility and hydrophobicity. Drug Discovery Today 2010, 15, (15/16), 648-655. 3. Leeson, P. D.; Springthorpe, B., The influence of drug-like concepts on decision- making in medicinal chemistry. Nature Reviews Drug Discovery 2007, 6, (11), 881-890. • Structural alerts in HTS 1. Baell, J. B.; Holloway, G. A., New substructure filters for removal of Pan Assay Interference Compounds (PAINS) from screening libraries and for their exclusion in bioassays. Journal of Medicinal Chemistry 2010, 53, (7), 2719-2740. 2. Rishton, G. M., Reactive compounds and in vitro false positives in HTS. Drug Discovery Today 1997, 2, (9), 382-384. 58 05/07/2012 Resources for Computa5onal Drug Design
- 59. Further reading • Similarity and diversity 1. Ashton, M.; Barnard, J.; Casset, F.; Charlton, M.; Downs, G.; Gorse, D.; Holliday, J.; Lahana, R.; Willett, P., Identification of diverse database subsets using property-based and fragment-based molecular descriptions. Quantitative Structure-Activity Relationships 2002, 21, (6), 598-604. 2. Bender, A.; Glen, R. C., Molecular similarity: a key technique in molecular informatics. Organic and Biomolecular Chemistry 2004, 2, 3204-3218. 3. Gorse, A.-D., Diversity in medicinal chemistry space. Current Topics in Medicinal Chemistry 2006, 6, (1), 3-18. 4. Maldonado, A.; Doucet, J.; Petitjean, M.; Fan, B.-T., Molecular similarity and diversity in chemoinformatics: From theory to applications. Molecular Diversity 2006, 10, (1), 39-79. 5. Rogers, D.; Hahn, M., Extended-connectivity fingerprints. Journal of Chemical Information and Modeling 2010, 50, (5), 742-754. 6. Schuffenhauer, A.; Brown, N., Chemical diversity and biological activity. Drug Discovery Today: Technologies 2006, 3, (4), 387-395. 7. Willett, P.; Barnard, J. M.; Downs, G. M., Chemical similarity searching. Journal of Chemical Information and Computer Sciences 1998, 38, (6), 983-996. 59 05/07/2012 Resources for Computa5onal Drug Design
- 60. Day 4: KNIME Tutorial George Papadatos, PhD Francis Atkinson, PhD ChEMBL group