Regression and Classification with R
- 2. cation with R Yanchang Zhao http://www.RDataMining.com 30 September 2014 1 / 44
- 3. Outline Introduction Linear Regression Generalized Linear Regression Decision Trees with Package party Decision Trees with Package rpart Random Forest Online Resources 2 / 44
- 5. cation with R 1 I build a linear regression model to predict CPI data I build a generalized linear model (GLM) I build decision trees with package party and rpart I train a random forest model with package randomForest 1Chapter 4: Decision Trees and Random Forest & Chapter 5: Regression, in book R and Data Mining: Examples and Case Studies. http://www.rdatamining.com/docs/RDataMining.pdf 3 / 44
- 6. Regression I Regression is to build a function of independent variables (also known as predictors) to predict a dependent variable (also called response). I For example, banks assess the risk of home-loan applicants based on their age, income, expenses, occupation, number of dependents, total credit limit, etc. I linear regression models I generalized linear models (GLM) 4 / 44
- 7. Outline Introduction Linear Regression Generalized Linear Regression Decision Trees with Package party Decision Trees with Package rpart Random Forest Online Resources 5 / 44
- 8. Linear Regression I Linear regression is to predict response with a linear function of predictors as follows: y = c0 + c1x1 + c2x2 + + ckxk ; where x1; x2; ; xk are predictors and y is the response to predict. I linear regression with function lm() I the Australian CPI (Consumer Price Index) data: quarterly CPIs from 2008 to 2010 2 2From Australian Bureau of Statistics, http://www.abs.gov.au. 6 / 44
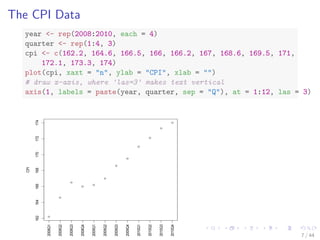
- 9. The CPI Data year - rep(2008:2010, each = 4) quarter - rep(1:4, 3) cpi - c(162.2, 164.6, 166.5, 166, 166.2, 167, 168.6, 169.5, 171, 172.1, 173.3, 174) plot(cpi, xaxt = n, ylab = CPI, xlab = ) # draw x-axis, where 'las=3' makes text vertical axis(1, labels = paste(year, quarter, sep = Q), at = 1:12, las = 3) 162 164 166 168 170 172 174 CPI 2008Q1 2008Q2 2008Q3 2008Q4 2009Q1 2009Q2 2009Q3 2009Q4 2010Q1 2010Q2 2010Q3 2010Q4 7 / 44
- 10. Linear Regression ## correlation between CPI and year / quarter cor(year, cpi) ## [1] 0.9096 cor(quarter, cpi) ## [1] 0.3738 ## build a linear regression model with function lm() fit - lm(cpi ~ year + quarter) fit ## ## Call: ## lm(formula = cpi ~ year + quarter) ## ## Coefficients: ## (Intercept) year quarter ## -7644.49 3.89 1.17 8 / 44
- 11. With the above linear model, CPI is calculated as cpi = c0 + c1 year + c2 quarter; where c0, c1 and c2 are coecients from model fit. What will the CPI be in 2011? cpi2011 - fit$coefficients[[1]] + fit$coefficients[[2]] * 2011 + fit$coefficients[[3]] * (1:4) cpi2011 ## [1] 174.4 175.6 176.8 177.9 9 / 44
- 12. With the above linear model, CPI is calculated as cpi = c0 + c1 year + c2 quarter; where c0, c1 and c2 are coecients from model fit. What will the CPI be in 2011? cpi2011 - fit$coefficients[[1]] + fit$coefficients[[2]] * 2011 + fit$coefficients[[3]] * (1:4) cpi2011 ## [1] 174.4 175.6 176.8 177.9 An easier way is to use function predict(). 9 / 44
- 13. More details of the model can be obtained with the code below. attributes(fit) ## $names ## [1] coefficients residuals effects ## [4] rank fitted.values assign ## [7] qr df.residual xlevels ## [10] call terms model ## ## $class ## [1] lm fit$coefficients ## (Intercept) year quarter ## -7644.488 3.888 1.167 10 / 44
- 14. Function residuals(): dierences between observed values and
- 15. tted values # differences between observed values and fitted values residuals(fit) ## 1 2 3 4 5 6 ... ## -0.57917 0.65417 1.38750 -0.27917 -0.46667 -0.83333 -0.40... ## 8 9 10 11 12 ## -0.66667 0.44583 0.37917 0.41250 -0.05417 summary(fit) ## ## Call: ## lm(formula = cpi ~ year + quarter) ## ## Residuals: ## Min 1Q Median 3Q Max ## -0.833 -0.495 -0.167 0.421 1.387 ## ## Coefficients: ## Estimate Std. Error t value Pr(|t|) ## (Intercept) -7644.488 518.654 -14.74 1.3e-07 *** ## year 3.888 0.258 15.06 1.1e-07 *** 11 / 44
- 16. 3D Plot of the Fitted Model library(scatterplot3d) s3d - scatterplot3d(year, quarter, cpi, highlight.3d = T, type = h, lab = c(2, 3)) # lab: number of tickmarks on x-/y-axes s3d$plane3d(fit) # draws the fitted plane 160 165 170 175 2008 2009 2010 1 2 3 4 year quarter cpi 12 / 44
- 17. Prediction of CPIs in 2011 data2011 - data.frame(year = 2011, quarter = 1:4) cpi2011 - predict(fit, newdata = data2011) style - c(rep(1, 12), rep(2, 4)) plot(c(cpi, cpi2011), xaxt = n, ylab = CPI, xlab = , pch = style, col = style) axis(1, at = 1:16, las = 3, labels = c(paste(year, quarter, sep = Q), 2011Q1, 2011Q2, 2011Q3, 2011Q4)) 165 170 175 CPI 2008Q1 2008Q2 2008Q3 2008Q4 2009Q1 2009Q2 2009Q3 2009Q4 2010Q1 2010Q2 2010Q3 2010Q4 2011Q1 2011Q2 2011Q3 2011Q4 13 / 44
- 18. Outline Introduction Linear Regression Generalized Linear Regression Decision Trees with Package party Decision Trees with Package rpart Random Forest Online Resources 14 / 44
- 19. Generalized Linear Model (GLM) I Generalizes linear regression by allowing the linear model to be related to the response variable via a link function and allowing the magnitude of the variance of each measurement to be a function of its predicted value I Uni
- 20. es various other statistical models, including linear regression, logistic regression and Poisson regression I Function glm():
- 21. ts generalized linear models, speci
- 22. ed by giving a symbolic description of the linear predictor and a description of the error distribution 15 / 44
- 23. Build a Generalized Linear Model data(bodyfat, package=TH.data) myFormula - DEXfat ~ age + waistcirc + hipcirc + elbowbreadth + kneebreadth bodyfat.glm - glm(myFormula, family = gaussian(log), data = bodyfat) summary(bodyfat.glm) ## ## Call: ## glm(formula = myFormula, family = gaussian(log), data = b... ## ## Deviance Residuals: ## Min 1Q Median 3Q Max ## -11.569 -3.006 0.127 2.831 10.097 ## ## Coefficients: ## Estimate Std. Error t value Pr(|t|) ## (Intercept) 0.73429 0.30895 2.38 0.0204 * ## age 0.00213 0.00145 1.47 0.1456 ## waistcirc 0.01049 0.00248 4.23 7.4e-05 *** ## hipcirc 0.00970 0.00323 3.00 0.0038 ** ## elbowbreadth 0.00235 0.04569 0.05 0.9590 ## kneebreadth 0.06319 0.02819 2.24 0.0284 * 16 / 44
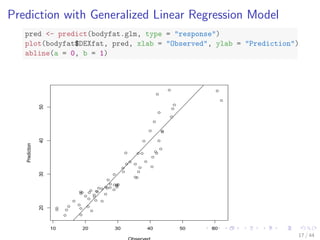
- 24. Prediction with Generalized Linear Regression Model pred - predict(bodyfat.glm, type = response) plot(bodyfat$DEXfat, pred, xlab = Observed, ylab = Prediction) abline(a = 0, b = 1) 10 20 30 40 50 60 20 30 40 50 Observed Prediction 17 / 44
- 25. Outline Introduction Linear Regression Generalized Linear Regression Decision Trees with Package party Decision Trees with Package rpart Random Forest Online Resources 18 / 44
- 26. The iris Data str(iris) ## 'data.frame': 150 obs. of 5 variables: ## $ Sepal.Length: num 5.1 4.9 4.7 4.6 5 5.4 4.6 5 4.4 4.9 ... ## $ Sepal.Width : num 3.5 3 3.2 3.1 3.6 3.9 3.4 3.4 2.9 3.1... ## $ Petal.Length: num 1.4 1.4 1.3 1.5 1.4 1.7 1.4 1.5 1.4 1... ## $ Petal.Width : num 0.2 0.2 0.2 0.2 0.2 0.4 0.3 0.2 0.2 0... ## $ Species : Factor w/ 3 levels setosa,versicolor,.... # split data into two subsets: training (70%) and test (30%); set # a fixed random seed to make results reproducible set.seed(1234) ind - sample(2, nrow(iris), replace = TRUE, prob = c(0.7, 0.3)) train.data - iris[ind == 1, ] test.data - iris[ind == 2, ] 19 / 44
- 27. Build a ctree I Control the training of decision trees: MinSplit, MinBusket, MaxSurrogate and MaxDepth I Target variable: Species I Independent variables: all other variables library(party) myFormula - Species ~ Sepal.Length + Sepal.Width + Petal.Length + Petal.Width iris_ctree - ctree(myFormula, data = train.data) # check the prediction table(predict(iris_ctree), train.data$Species) ## ## setosa versicolor virginica ## setosa 40 0 0 ## versicolor 0 37 3 ## virginica 0 1 31 20 / 44
- 28. Print ctree print(iris_ctree) ## ## Conditional inference tree with 4 terminal nodes ## ## Response: Species ## Inputs: Sepal.Length, Sepal.Width, Petal.Length, Petal.Width ## Number of observations: 112 ## ## 1) Petal.Length = 1.9; criterion = 1, statistic = 104.643 ## 2)* weights = 40 ## 1) Petal.Length 1.9 ## 3) Petal.Width = 1.7; criterion = 1, statistic = 48.939 ## 4) Petal.Length = 4.4; criterion = 0.974, statistic = ... ## 5)* weights = 21 ## 4) Petal.Length 4.4 ## 6)* weights = 19 ## 3) Petal.Width 1.7 ## 7)* weights = 32 21 / 44
- 29. plot(iris_ctree) 1 Petal.Length p 0.001 £ 1.9 1.9 Node 2 (n = 40) setosa 1 0.8 0.6 0.4 0.2 0 3 Petal.Width p 0.001 £ 1.7 1.7 4 Petal.Length p = 0.026 £ 4.4 4.4 Node 5 (n = 21) setosa 1 0.8 0.6 0.4 0.2 0 Node 6 (n = 19) setosa 1 0.8 0.6 0.4 0.2 0 Node 7 (n = 32) setosa 1 0.8 0.6 0.4 0.2 0 22 / 44
- 30. plot(iris_ctree, type = simple) 1 Petal.Length p 0.001 £ 1.9 1.9 2 n = 40 y = (1, 0, 0) 3 Petal.Width p 0.001 £ 1.7 1.7 4 Petal.Length p = 0.026 £ 4.4 4.4 5 n = 21 y = (0, 1, 0) 6 n = 19 y = (0, 0.842, 0.158) 7 n = 32 y = (0, 0.031, 0.969) 23 / 44
- 31. Test # predict on test data testPred - predict(iris_ctree, newdata = test.data) table(testPred, test.data$Species) ## ## testPred setosa versicolor virginica ## setosa 10 0 0 ## versicolor 0 12 2 ## virginica 0 0 14 24 / 44
- 32. Outline Introduction Linear Regression Generalized Linear Regression Decision Trees with Package party Decision Trees with Package rpart Random Forest Online Resources 25 / 44
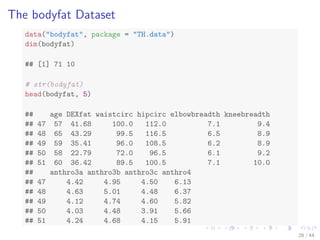
- 33. The bodyfat Dataset data(bodyfat, package = TH.data) dim(bodyfat) ## [1] 71 10 # str(bodyfat) head(bodyfat, 5) ## age DEXfat waistcirc hipcirc elbowbreadth kneebreadth ## 47 57 41.68 100.0 112.0 7.1 9.4 ## 48 65 43.29 99.5 116.5 6.5 8.9 ## 49 59 35.41 96.0 108.5 6.2 8.9 ## 50 58 22.79 72.0 96.5 6.1 9.2 ## 51 60 36.42 89.5 100.5 7.1 10.0 ## anthro3a anthro3b anthro3c anthro4 ## 47 4.42 4.95 4.50 6.13 ## 48 4.63 5.01 4.48 6.37 ## 49 4.12 4.74 4.60 5.82 ## 50 4.03 4.48 3.91 5.66 ## 51 4.24 4.68 4.15 5.91 26 / 44
- 34. Train a Decision Tree with Package rpart # split into training and test subsets set.seed(1234) ind - sample(2, nrow(bodyfat), replace=TRUE, prob=c(0.7, 0.3)) bodyfat.train - bodyfat[ind==1,] bodyfat.test - bodyfat[ind==2,] # train a decision tree library(rpart) myFormula - DEXfat ~ age + waistcirc + hipcirc + elbowbreadth + kneebreadth bodyfat_rpart - rpart(myFormula, data = bodyfat.train, control = rpart.control(minsplit = 10)) # print(bodyfat_rpart$cptable) print(bodyfat_rpart) plot(bodyfat_rpart) text(bodyfat_rpart, use.n=T) 27 / 44
- 35. The rpart Tree ## n= 56 ## ## node), split, n, deviance, yval ## * denotes terminal node ## ## 1) root 56 7265.0000 30.95 ## 2) waistcirc 88.4 31 960.5000 22.56 ## 4) hipcirc 96.25 14 222.3000 18.41 ## 8) age 60.5 9 66.8800 16.19 * ## 9) age=60.5 5 31.2800 22.41 * ## 5) hipcirc=96.25 17 299.6000 25.97 ## 10) waistcirc 77.75 6 30.7300 22.32 * ## 11) waistcirc=77.75 11 145.7000 27.96 ## 22) hipcirc 99.5 3 0.2569 23.75 * ## 23) hipcirc=99.5 8 72.2900 29.54 * ## 3) waistcirc=88.4 25 1417.0000 41.35 ## 6) waistcirc 104.8 18 330.6000 38.09 ## 12) hipcirc 109.9 9 69.0000 34.38 * ## 13) hipcirc=109.9 9 13.0800 41.81 * ## 7) waistcirc=104.8 7 404.3000 49.73 * 28 / 44
- 36. The rpart Tree waistcir|c 88.4 hipcirc 96.25 age 60.5 waistcirc 77.75 hipcirc 99.5 waistcirc 104.8 2e+01 hipcirc 109.9 n=9 2e+01 n=5 2e+01 n=6 2e+01 n=3 3e+01 n=8 3e+01 n=9 4e+01 n=9 5e+01 n=7 29 / 44
- 37. Select the Best Tree # select the tree with the minimum prediction error opt - which.min(bodyfat_rpart$cptable[, xerror]) cp - bodyfat_rpart$cptable[opt, CP] # prune tree bodyfat_prune - prune(bodyfat_rpart, cp = cp) # plot tree plot(bodyfat_prune) text(bodyfat_prune, use.n = T) 30 / 44
- 38. Selected Tree waistcir|c 88.4 hipcirc 96.25 age 60.5 waistcirc 77.75 waistcirc 104.8 hipcirc 109.9 2e+01 n=9 2e+01 n=5 2e+01 n=6 3e+01 n=11 3e+01 n=9 4e+01 n=9 5e+01 n=7 31 / 44
- 39. Model Evalutation DEXfat_pred - predict(bodyfat_prune, newdata = bodyfat.test) xlim - range(bodyfat$DEXfat) plot(DEXfat_pred ~ DEXfat, data = bodyfat.test, xlab = Observed, ylab = Prediction, ylim = xlim, xlim = xlim) abline(a = 0, b = 1) 10 20 30 40 50 60 10 20 30 40 50 60 Observed Prediction 32 / 44
- 40. Outline Introduction Linear Regression Generalized Linear Regression Decision Trees with Package party Decision Trees with Package rpart Random Forest Online Resources 33 / 44
- 41. R Packages for Random Forest I Package randomForest I very fast I cannot handle data with missing values I a limit of 32 to the maximum number of levels of each categorical attribute I Package party: cforest() I not limited to the above maximum levels I slow I needs more memory 34 / 44
- 42. Train a Random Forest # split into two subsets: training (70%) and test (30%) ind - sample(2, nrow(iris), replace=TRUE, prob=c(0.7, 0.3)) train.data - iris[ind==1,] test.data - iris[ind==2,] # use all other variables to predict Species library(randomForest) rf - randomForest(Species ~ ., data=train.data, ntree=100, proximity=T) 35 / 44
- 43. table(predict(rf), train.data$Species) ## ## setosa versicolor virginica ## setosa 36 0 0 ## versicolor 0 31 2 ## virginica 0 1 34 print(rf) ## ## Call: ## randomForest(formula = Species ~ ., data = train.data, ntr... ## Type of random forest: classification ## Number of trees: 100 ## No. of variables tried at each split: 2 ## ## OOB estimate of error rate: 2.88% ## Confusion matrix: ## setosa versicolor virginica class.error ## setosa 36 0 0 0.00000 ## versicolor 0 31 1 0.03125 ## virginica 0 2 34 0.05556 36 / 44
- 44. Error Rate of Random Forest plot(rf, main = ) 0 20 40 60 80 100 0.00 0.05 0.10 0.15 0.20 trees Error 37 / 44
- 45. Variable Importance importance(rf) ## MeanDecreaseGini ## Sepal.Length 6.914 ## Sepal.Width 1.283 ## Petal.Length 26.267 ## Petal.Width 34.164 38 / 44
- 46. Variable Importance varImpPlot(rf) Petal.Width Petal.Length Sepal.Length Sepal.Width rf 0 5 10 15 20 25 30 35 MeanDecreaseGini 39 / 44
- 47. Margin of Predictions The margin of a data point is as the proportion of votes for the correct class minus maximum proportion of votes for other classes. Positive margin means correct classi
- 48. cation. irisPred - predict(rf, newdata = test.data) table(irisPred, test.data$Species) ## ## irisPred setosa versicolor virginica ## setosa 14 0 0 ## versicolor 0 17 3 ## virginica 0 1 11 plot(margin(rf, test.data$Species)) 40 / 44
- 49. Margin of Predictions 0 20 40 60 80 100 0.0 0.2 0.4 0.6 0.8 1.0 Index x 41 / 44
- 50. Outline Introduction Linear Regression Generalized Linear Regression Decision Trees with Package party Decision Trees with Package rpart Random Forest Online Resources 42 / 44
- 51. Online Resources I Chapter 4: Decision Trees and Random Forest Chapter 5: Regression, in book R and Data Mining: Examples and Case Studies http://www.rdatamining.com/docs/RDataMining.pdf I R Reference Card for Data Mining http://www.rdatamining.com/docs/R-refcard-data-mining.pdf I Free online courses and documents http://www.rdatamining.com/resources/ I RDataMining Group on LinkedIn (7,000+ members) http://group.rdatamining.com I RDataMining on Twitter (1,700+ followers) @RDataMining 43 / 44
- 52. The End Thanks! Email: yanchang(at)rdatamining.com 44 / 44








![Linear Regression
## correlation between CPI and year / quarter
cor(year, cpi)
## [1] 0.9096
cor(quarter, cpi)
## [1] 0.3738
## build a linear regression model with function lm()
fit - lm(cpi ~ year + quarter)
fit
##
## Call:
## lm(formula = cpi ~ year + quarter)
##
## Coefficients:
## (Intercept) year quarter
## -7644.49 3.89 1.17
8 / 44](https://image.slidesharecdn.com/rdatamining-slides-regression-classification-140915032901-phpapp01/85/Regression-and-Classification-with-R-10-320.jpg)
![With the above linear model, CPI is calculated as
cpi = c0 + c1 year + c2 quarter;
where c0, c1 and c2 are coecients from model fit.
What will the CPI be in 2011?
cpi2011 - fit$coefficients[[1]] +
fit$coefficients[[2]] * 2011 +
fit$coefficients[[3]] * (1:4)
cpi2011
## [1] 174.4 175.6 176.8 177.9
9 / 44](https://image.slidesharecdn.com/rdatamining-slides-regression-classification-140915032901-phpapp01/85/Regression-and-Classification-with-R-11-320.jpg)
![With the above linear model, CPI is calculated as
cpi = c0 + c1 year + c2 quarter;
where c0, c1 and c2 are coecients from model fit.
What will the CPI be in 2011?
cpi2011 - fit$coefficients[[1]] +
fit$coefficients[[2]] * 2011 +
fit$coefficients[[3]] * (1:4)
cpi2011
## [1] 174.4 175.6 176.8 177.9
An easier way is to use function predict().
9 / 44](https://image.slidesharecdn.com/rdatamining-slides-regression-classification-140915032901-phpapp01/85/Regression-and-Classification-with-R-12-320.jpg)
![More details of the model can be obtained with the code below.
attributes(fit)
## $names
## [1] coefficients residuals effects
## [4] rank fitted.values assign
## [7] qr df.residual xlevels
## [10] call terms model
##
## $class
## [1] lm
fit$coefficients
## (Intercept) year quarter
## -7644.488 3.888 1.167
10 / 44](https://image.slidesharecdn.com/rdatamining-slides-regression-classification-140915032901-phpapp01/85/Regression-and-Classification-with-R-13-320.jpg)











![The iris Data
str(iris)
## 'data.frame': 150 obs. of 5 variables:
## $ Sepal.Length: num 5.1 4.9 4.7 4.6 5 5.4 4.6 5 4.4 4.9 ...
## $ Sepal.Width : num 3.5 3 3.2 3.1 3.6 3.9 3.4 3.4 2.9 3.1...
## $ Petal.Length: num 1.4 1.4 1.3 1.5 1.4 1.7 1.4 1.5 1.4 1...
## $ Petal.Width : num 0.2 0.2 0.2 0.2 0.2 0.4 0.3 0.2 0.2 0...
## $ Species : Factor w/ 3 levels setosa,versicolor,....
# split data into two subsets: training (70%) and test (30%); set
# a fixed random seed to make results reproducible
set.seed(1234)
ind - sample(2, nrow(iris), replace = TRUE, prob = c(0.7, 0.3))
train.data - iris[ind == 1, ]
test.data - iris[ind == 2, ]
19 / 44](https://image.slidesharecdn.com/rdatamining-slides-regression-classification-140915032901-phpapp01/85/Regression-and-Classification-with-R-26-320.jpg)





![The bodyfat Dataset
data(bodyfat, package = TH.data)
dim(bodyfat)
## [1] 71 10
# str(bodyfat)
head(bodyfat, 5)
## age DEXfat waistcirc hipcirc elbowbreadth kneebreadth
## 47 57 41.68 100.0 112.0 7.1 9.4
## 48 65 43.29 99.5 116.5 6.5 8.9
## 49 59 35.41 96.0 108.5 6.2 8.9
## 50 58 22.79 72.0 96.5 6.1 9.2
## 51 60 36.42 89.5 100.5 7.1 10.0
## anthro3a anthro3b anthro3c anthro4
## 47 4.42 4.95 4.50 6.13
## 48 4.63 5.01 4.48 6.37
## 49 4.12 4.74 4.60 5.82
## 50 4.03 4.48 3.91 5.66
## 51 4.24 4.68 4.15 5.91
26 / 44](https://image.slidesharecdn.com/rdatamining-slides-regression-classification-140915032901-phpapp01/85/Regression-and-Classification-with-R-33-320.jpg)
![Train a Decision Tree with Package rpart
# split into training and test subsets
set.seed(1234)
ind - sample(2, nrow(bodyfat), replace=TRUE, prob=c(0.7, 0.3))
bodyfat.train - bodyfat[ind==1,]
bodyfat.test - bodyfat[ind==2,]
# train a decision tree
library(rpart)
myFormula - DEXfat ~ age + waistcirc + hipcirc + elbowbreadth +
kneebreadth
bodyfat_rpart - rpart(myFormula, data = bodyfat.train,
control = rpart.control(minsplit = 10))
# print(bodyfat_rpart$cptable)
print(bodyfat_rpart)
plot(bodyfat_rpart)
text(bodyfat_rpart, use.n=T)
27 / 44](https://image.slidesharecdn.com/rdatamining-slides-regression-classification-140915032901-phpapp01/85/Regression-and-Classification-with-R-34-320.jpg)


![Select the Best Tree
# select the tree with the minimum prediction error
opt - which.min(bodyfat_rpart$cptable[, xerror])
cp - bodyfat_rpart$cptable[opt, CP]
# prune tree
bodyfat_prune - prune(bodyfat_rpart, cp = cp)
# plot tree
plot(bodyfat_prune)
text(bodyfat_prune, use.n = T)
30 / 44](https://image.slidesharecdn.com/rdatamining-slides-regression-classification-140915032901-phpapp01/85/Regression-and-Classification-with-R-37-320.jpg)




![Train a Random Forest
# split into two subsets: training (70%) and test (30%)
ind - sample(2, nrow(iris), replace=TRUE, prob=c(0.7, 0.3))
train.data - iris[ind==1,]
test.data - iris[ind==2,]
# use all other variables to predict Species
library(randomForest)
rf - randomForest(Species ~ ., data=train.data, ntree=100,
proximity=T)
35 / 44](https://image.slidesharecdn.com/rdatamining-slides-regression-classification-140915032901-phpapp01/85/Regression-and-Classification-with-R-42-320.jpg)


