Scalable Parallel Programming in Python with Parsl
- 1. Scalable Parallel Programming in Python with Parsl Kyle Chard ([email protected]) Yadu Babuji, Anna Woodard, Ben Clifford, Zhuozhao Li, Mike Wilde, Dan Katz, Ian Foster http://parsl-project.org
- 2. 2 Composition and parallelism Scientific software is increasingly assembled rather than written – High-level language to integrate and wrap components from many sources Parallel and distributed computing is ubiquitous – Increasing data sizes combined with plateauing sequential processing power Python (and the SciPy ecosystem) is the de facto standard language for science – Libraries, tools, Jupyter, etc. Parsl allows for the natural expression of parallelism in Python: – Programs can express opportunities for parallelism – Realized, at execution time, using different execution models on different parallel platforms
- 3. 3 Parsl: parallel programming in Python Apps define opportunities for parallelism • Python apps call Python functions • Bash apps call external applications Apps return “futures”: a proxy for a result that might not yet be available Apps run concurrently respecting data dependencies. Natural parallel programming! Parsl scripts are independent of where they run. Write once run anywhere! pip install parsl Try Parsl: https://mybinder.org/v2/gh/Parsl/parsl-tutorial/master
- 4. 4 Data-driven example: parallel geospatial analysis Land-use Image processing pipeline for the MODIS remote sensor Analyze Landuse Colorize Mark Assemble
- 5. 5 Expressing a many task workflow in Parsl 1) Wrap the science applications as Parsl Apps: @bash_app def simulate(outputs=[]): return './simulation_app.exe {outputs[0]}’ @bash_app def merge(inputs=[], outputs=[]): i = inputs; o = outputs return './merge {1} {0}'.format(' '.join(i), o[0]) @python_app def analyze(inputs=[]): return analysis_package(inputs)
- 6. 6 Expressing a many task workflow in Parsl 2) Execute the parallel workflow by calling Apps: sims = [] for i in range (nsims): sims.append(simulate(outputs=['sim-%s.txt' % i])) all = merge(inputs=[i.outputs[0] for i in sims], outputs=['all.txt']) result = analyze(inputs=[all.outputs[0]])
- 7. 7 Decomposing dynamic parallel execution into a task- dependency graph Parsl
- 8. 8 Parsl scripts are execution provider independent The same script can be run locally, on grids, clouds, or supercomputers Growing support for various schedulers and cloud vendors
- 9. 9 Separation of code and execution Choose execution environment at runtime. Parsl will direct tasks to the configured execution environment(s).
- 10. 10 Authentication and authorization Authn/z is hard… – 2FA, X509, GSISSH, etc. Integration with Globus Auth to support native app integration for accessing Globus (and other) services Using scoped access tokens, refresh tokens, delegation support
- 11. 11 Parsl provides transparent (wide area) data management Implicit data movement to/from repositories, laptops, supercomputers Globus for third-party, high performance and reliable data transfer – Support for site-specific DTNs HTTP/FTP direct data staging parsl_file = File(globus://EP/path/file) www.globus.org
- 12. 12 Parallel applications are very different High-throughput workloads – Protein docking, image processing, materials reconstructions – Requirements: 1000s of tasks, 100s of nodes, reliability, usability, monitoring, elasticity, etc. Extreme-scale workloads – Cosmology simulations, imaging the arctic, genomics analysis – Requirements: millions of tasks, 1000s of nodes (100,000s cores), capacity Interactive and real-time workloads – Materials science, cosmic ray shower analysis, machine learning inference – Requirements: 10s of nodes, rapid response, pipelining
- 13. 13 Parsl implements an extensible executor interface High-throughput executor (HTEX) – Pilot job-based model with multi-threaded manager deployed on workers – Designed for ease of use, fault-tolerance, etc. – <2000 nodes (~60K workers), Ms tasks, task duration/nodes > 0.01 Extreme-scale executor (EXEX) – Distributed MPI job manages execution. Manager rank communicates workload to other worker ranks directly – Designed for extreme scale execution on supercomputers – >1000 nodes (>30K workers), Ms tasks, >1m task duration Low-latency Executor (LLEX) – Direct socket communication to workers, fixed resource pool, limited features – 10s nodes, <1M tasks, <1m tasks
- 14. 14 Parsl executors scale to 2M tasks/256K workers Weak scaling: 10 tasks per worker ● HTEX and EXEX outperform other Python-based approaches and scale beyond ~2M tasks ● HTEX and EXEX scale to 2K nodes (~65k workers) and 8K nodes (~262K workers), respectively, with >1K tasks/s 0s tasks 1s tasks
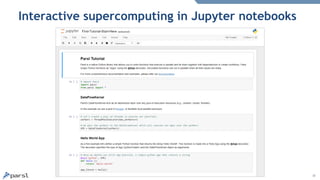
- 15. 15 Interactive supercomputing in Jupyter notebooks
- 17. 17 Other functionality provided by Parsl Globus. Delegated authentication and wide area data management Fault tolerance. Support for retries, checkpointing, and memoization Containers. Sandboxed execution environments for workers and tasks Data management. Automated staging with HTTP, FTP, and Globus Multi site. Combining executors/providers for execution across different resources Elasticity. Automated resource expansion/retraction based on workload Monitoring. Workflow and resource monitoring and visualization Reproducibility. Capture workflow provenance in the task graph Jupyter integration. Seamless description and management of workflows Resource abstraction. Block-based model overlaying different providers and resources
- 18. 18 Parsl is being used in a wide range of scientific applications E C A B D G • Machine learning to predict stopping power in materials • Protein and biomolecule structure and interaction • Weak lensing using sky surveys • Cosmic ray showers as part of QuarkNet • Information extraction to classify image types in papers • Materials science at the Advanced Photon Source • Machine learning and data analytics (DLHub) A B C D E F G F
- 19. 19 Parsl provides simple, safe, scalable, and flexible parallelism in Python Simple: Python with minimal new constructs (integrated with the growing SciPy ecosystem and other scientific services) Safe: deterministic parallel programs through immutable input/output objects, dependency task graph, etc. Scalable: efficient execution from laptops to the largest supercomputers Flexible: programs composed from existing components and then applied to different resources/workloads
- 20. 20 Parsl is an open-source Python project https://github.com/Parsl/parsl
- 21. 21 Questions? U . S . D E P A R T M E N T O F ENERGY http://parsl-project.org https://mybinder.org/v2/gh/Parsl/parsl-tutorial/master





![5
Expressing a many task workflow in Parsl
1) Wrap the science applications as Parsl Apps:
@bash_app
def simulate(outputs=[]):
return './simulation_app.exe {outputs[0]}’
@bash_app
def merge(inputs=[], outputs=[]):
i = inputs; o = outputs
return './merge {1} {0}'.format(' '.join(i), o[0])
@python_app
def analyze(inputs=[]):
return analysis_package(inputs)](https://image.slidesharecdn.com/19-7parsl-pearc-190807182457/85/Scalable-Parallel-Programming-in-Python-with-Parsl-5-320.jpg)
![6
Expressing a many task workflow in Parsl
2) Execute the parallel workflow by calling Apps:
sims = []
for i in range (nsims):
sims.append(simulate(outputs=['sim-%s.txt' % i]))
all = merge(inputs=[i.outputs[0] for i in sims],
outputs=['all.txt'])
result = analyze(inputs=[all.outputs[0]])](https://image.slidesharecdn.com/19-7parsl-pearc-190807182457/85/Scalable-Parallel-Programming-in-Python-with-Parsl-6-320.jpg)