Use of semantic phenotyping to aid disease diagnosis
- 1. Use of semantic phenotyping to aid disease diagnosis Melissa Haendel July 10th, 2014
- 2. Outline Semantic Diagnosis of known diseases Semantic similarity across species Combining Exome analysis with cross- species semantic phenotyping How much phenotyping is enough?
- 3. The undiagnosed patient Known disorders not recognized during prior evaluations? Atypical presentation of known disorders? Combinations of several disorders? Novel, unreported disorder?
- 4. OMIM Query # Records “large bone” 785 “enlarged bone” 156 “big bone” 16 “huge bones” 4 “massive bones” 28 “hyperplastic bones” 12 “hyperplastic bone” 40 “bone hyperplasia” 134 “increased bone growth” 612 Searching for phenotypes using text alone is insufficient
- 5. The Challenge: Interpretation of Disease Candidates ? What’s in the box? How are candidates identified? How do they compare? Prioritized Candidates, Models, functional validation M1 M2 M3 M4 ... Phenotypes P1 P2 P3 … Genotype info G1 G2 G3 G4 … Pathogenicity, frequency, protein interactions, gene expression, gene networks, epigenomics, metabolomics….
- 6. What is an ontology? A set of logically defined, inter-related terms used to annotate data Use of common or logically related terms across databases enables integration Relationships between terms allow annotations to be grouped in scientifically meaningful ways Reasoning software enables computation of inferred knowledge Groups of annotations can be compared using semantic similarity algorithms
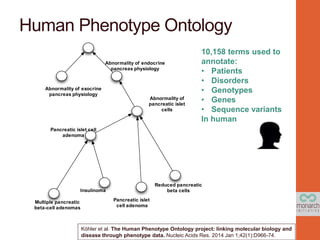
- 7. Human Phenotype Ontology 10,158 terms used to annotate: • Patients • Disorders • Genotypes • Genes • Sequence variants In human Reduced pancreatic beta cells Abnormality of pancreatic islet cells Abnormality of endocrine pancreas physiology Pancreatic islet cell adenoma Pancreatic islet cell adenoma Insulinoma Multiple pancreatic beta-cell adenomas Abnormality of exocrine pancreas physiology Köhler et al. The Human Phenotype Ontology project: linking molecular biology and disease through phenotype data. Nucleic Acids Res. 2014 Jan 1;42(1):D966-74.
- 8. A human phenotype example Abnormality of the eye Vitreous hemorrhage Abnormal eye morphology Abnormality of the cardiovascular system Abnormal eye physiology Hemorrhage of the eye Internal hemorrhage Abnormality of the globe Abnormality of blood circulation
- 9. ➔Phenotype annotations are unevenly distributed across different anatomical systems Survey of Annotations in Disease Corpus 7,401 diseases 99,045 annotations
- 10. exome analysis Recessive, De novo filters Remove off-target, common variants, and variants not in known disease causing genes Zemojtelet al., manuscript in presshttp://compbio.charite.de/PhenIX/ Target panel of 2,742 known Mendelian disease genes Compare phenotype profiles using data from: HGMD, Clinvar, OMIM, Orphanet
- 11. PhenIX performance testing Simulated datasets for a given disease and inheritance model created by spiking DAG panel generated VCF file with mutations from HGMD
- 12. PhenIX helped diagnose 11/38 patients global developmental delay (HP:0001263) delayed speech and language development (HP:0000750) motor delay (HP:0001270) proportionate short stature (HP:0003508) microcephaly (HP:0000252) feeding difficulties (HP:0011968) congenital megaloureter(HP:0008676) cone-shaped epiphysis of the phalanges of the hand (HP:0010230) sacral dimple (HP:0000960) hyperpigmentated/hypopigmentated macules (HP:0007441) hypertelorism (HP:0000316) abnormality of the midface (HP:0000309) flat nose (HP:0000457) thick lower lip vermilion (HP:0000179) thick upper lip vermilion (HP:0000215) full cheeks (HP:0000293) short neck (HP:0000470)
- 13. What to do when we can’t diagnose with a known disease?
- 14. Outline Semantic Diagnosis of known diseases Semantic similarity across species Combining Exome analysis with cross- species semantic phenotyping How much phenotyping is enough?
- 15. B6.Cg-Alms1foz/fox/J increased weight, adipose tissue volume, glucose homeostasis altered ALSM1(NM_015120.4) [c.10775delC] + [-] GENOTYPE PHENOTYPE obesity, diabetes mellitus, insulin resistance increased food intake, hyperglycemia, insulin resistance kcnj11c14/c14; insrt143/+(AB) Models recapitulate various phenotypic aspects of disease ?
- 16. How much phenotype data? • Human genes have poor phenotype coverage GWAS + ClinVar + OMIM
- 17. How much phenotype data? • Human genes have poor phenotype coverage • What else can we leverage? GWAS + ClinVar + OMIM
- 18. How much phenotype data? • Human genes have poor phenotype coverage • What else can we leverage? …animal models Orthology via PANTHER v9
- 19. How much phenotype data? • Combined, human and model phenotypes can be linked to >75% human genes. Orthology via PANTHER v9
- 20. Monarch phenotype data Also in the system: Rat; IMPC; GO annotations; Coriell cell lines; OMIA; MPD; Yeast; CTD; GWAS; Panther, Homologene orthologs; BioGrid interactions; Drugbank; AutDB; Allen Brain …157 sources to date Coming soon: Animal QTLs for pig, cattle, chicken, sheep, trout, dog, horse Species Data source Genes Genotypes Variants Phenotype annotations Diseases mouse MGI 13,433 59,087 34,895 271,621 fish ZFIN 7,612 25,588 17,244 81,406 fly Flybase 27,951 91,096 108,348 267,900 worm Wormbase 23,379 15,796 10,944 543,874 human HPOA 112,602 7,401 human OMIM 2,970 4,437 3,651 human ClinVar 3,215 100,523 445,241 4,056 human KEGG 2,509 3,927 1,159 human ORPHANET 3,113 5,690 3,064 human CTD 7,414 23,320 4,912
- 21. Survey of Annotations Disease/Model Corpus Data from MGI, ZFIN, & HPO, reasoned over with cross-species phenotype ontology https://code.google.com/p/phenotype-ontologies/ ➔Models have a different phenotype distribution
- 22. Multiple ways to compare disease to models Asserted models Inferred by orthology Inferred by gene enrichment Inferred by phenotypic similarity
- 23. Models based on phenotypic similarity Washington, N. L., Haendel, M. A., Mungall, C. J., Ashburner, M., Westerfield, M., & Lewis, S. E. (2009). Linking Human Diseases to Animal Models Using Ontology-Based Phenotype Annotation. PLoS Biol, 7(11). doi:10.1371/journal.pbio.1000247
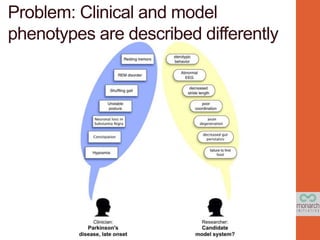
- 24. Problem: Clinical and model phenotypes are described differently
- 25. lung lung lobular organ parenchymatous organ solid organ pleural sac thoracic cavity organ thoracic cavity abnormal lung morphology abnormal respiratory system morphology Mammalian Phenotype Mouse Anatomy FMA abnormal pulmonary acinus morphology abnormal pulmonary alveolus morphology lung alveolus organ system respiratory system Lower respiratory tract alveolar sac pulmonary acinus organ system respiratory system Human development lung lung bud respiratory primordium pharyngeal region Another Problem: Data silos develops_from part_of is_a (SubClassOf) surrounded_by
- 26. Solution: bridging semantics Mungall, C. J., Torniai, C., Gkoutos, G. V., Lewis, S. E., & Haendel, M. A. (2012). Uberon, an integrative multi-species anatomy ontology. Genome Biology, 13(1), R5. doi:10.1186/gb-2012-13-1-r5 anatomical structure endoderm of forgut lung bud lung respiration organ organ foregut alveolus alveolus of lung organ part FMA:lung MA:lung endoderm GO: respiratory gaseous exchange MA:lung alveolus FMA: pulmonary alveolus is_a (taxon equivalent) develops_from part_of is_a (SubClassOf) capable_of NCBITaxon: Mammalia EHDAA: lung bud only_in_taxon pulmonary acinus alveolar sac lung primordium swim bladder respiratory primordium NCBITaxon: Actinopterygii Haendel, M. A. et al. (2014). Unification of multi-species vertebrate anatomy ontologies for comparative biology in Uberon. Journal of Biomedical Semantics 2014, 5:21. doi:10.1186/2041-1480-5-21
- 27. Modular phenotype description Entity (Anatomy, Spatial, Gene Ontology) BSPO: anterior region part_of ZFA:head ZFA:heart ZFA:ventral mandibular arch GO:swim bladder inflation Quality (PATO) Small size Edematous Thick Arrested
- 28. Mammalian Phenotype Ontology Smith et al. (2005). The Mammalian Phenotype Ontology as a tool for annotating, analyzing and comparing phenotypic information. Genome Biol, 6(1). doi:10.1186/gb-2004-6-1-r7 10,097 terms used to annotate and query: • Genotypes • Alleles • Genes In mice abnormal pancreatic beta cell mass abnormal pancreatic beta cell morphology abnormal pancreatic islet morphology abnormal endocrine pancreas morphology abnormal pancreatic beta cell differentiation abnormal pancreatic alpha cell morphology abnormal pancreatic alpha cell differentiation abnormal pancreatic alpha cell number
- 29. Phenotype representation requires more than “phenotype ontologies” glucose metabolism (GO:0006006) Gene/protein function data glucose (CHEBI:172 34) Metabolomics, toxicogenomics data Disease & phenotype data type II diabetes mellitus (DOID:9352) pyruvate (CHEBI:153 61) Disease Gene Ontology Chemical pancreatic beta cell (CL:0000169) transcriptomic data Cell
- 30. Uberpheno – building a cross- species semantic framework Köhler et al. (2014) Construction and accessibility of a cross-species phenotype ontology along with gene annotations for biomedical research F1000Research 2014, 2:30
- 35. OWLsim: Phenotype similarity across patients or organisms Unstable posture Constipation Neuronal loss in Substantia Nigra Shuffling gait Resting tremors REM disorder Hyposmia poor rotarod performance decreased gut peristalsis axon degeneration decreased stride length sterotypic behavior abnormal EEG failure to find food abnormal coordination abnormal digestive physiology CNS neuron degeneration abnormal locomotion abnormal motor function sleep disturbance abnormal olfaction https://code.google.com/p/owltools/wiki/OwlSim
- 36. Visualizing phenotypic similarity ➔Each model recapitulates some of the disease phenotypes Holoprosencephaly I (unknown gene, mapped to 21q22.3) compared to most similar mouse models
- 37. Models of disease based on phenotypic similarity Holoprosencephaly I (unknown gene, mapped to 21q22.3) compared to most similar mouse models ➔The ontologies enable comparison across species
- 38. Outline Semantic Diagnosis of known diseases Semantic similarity across species Combining Exome analysis with cross- species semantic phenotyping How much phenotyping is enough?
- 40. Exomiser results for the Undiagnosed Disease Program 11 previously diagnosed families Exomiser 2.0 identified the causative variants with a rank of at least 7/408 potential variants 23 families without identified disorders We have now prioritized variants in STIM1, ATP13A2, PANK2, and CSF1R in 5 different families (2 STIM1 families)
- 41. Exomiser performance on solved UDP cases 0 1 2 3 4 5 6 7 8 9 10 11 Exo Variant Exo Pheno Exo Exo no Mendelian Exo Novel top10 top 5 top candidate
- 42. UDP_2731 candidates Chromosome Position Reference Allele Variant Allele GENE Phenotype score Variant Score Exomiser Score chrX 19554576T C SH3KBP1 0.5051473 0.995576 0.7503617 chr2 179658310T C TTN 0.64627105 0.79311335 0.71969223 chr2 179632598C T TTN 0.64627105 0.79311335 0.71969223 chr2 179567340G A TTN 0.64627105 0.79311335 0.71969223 chr2 179553542G T TTN 0.64627105 0.79311335 0.71969223 chr2 179549131C T TTN 0.64627105 0.79311335 0.71969223 chr18 67836115G T RTTN 0.7629328 0.25979215 0.51136243 chr18 67721492G C RTTN 0.7629328 0.25979215 0.51136243 chr18 67673764T C RTTN 0.7629328 0.25979215 0.51136243 chrX 140993905- GCTCCTTCTCCTCCACTTTATTGAG TATTTTCCAGAGTTCCCCTGAGAG AAGTCAGAGAACTTCTGAGGGTTT TGCACAGTCTCCTCTCCAGATTCCT GTGAGCT MAGEC1 0.5416666 0.85 0.6958333 chr6 30858858G A DDR1 0.37619072 1 0.68809533 chr3 129308149 AGCCTCCCACCCCCACCCCCT CCCCACATCCCCAACCATACC TACCTTGAGA - PLXND1 0.34432834 0.95 0.64716417 chr5 37245866G A C5orf42 0.7855199 0.5 0.6427599 chr5 37169169T C C5orf42 0.7855199 0.5 0.6427599 chr6 42946264G A PEX6 0.7187602 0.5 0.6093801 chr6 42931861G A PEX6 0.7187602 0.5 0.6093801 chrX 53113897G C TSPYL2 0.59999996 0.4906897 0.5453448 chr13 75911097T C TBC1D4 0.23643239 0.7895149 0.51297367 chr13 75900510G A TBC1D4 0.23643239 0.7895149 0.51297367 chr13 75861174- A TBC1D4 0.23643239 0.7895149 0.51297367 chr18 67836115G T RTTN 0.7629328 0.25979215 0.51136243 chr18 67721492G C RTTN 0.7629328 0.25979215 0.51136243 chr18 67673764T C RTTN 0.7629328 0.25979215 0.51136243
- 43. UDP_2731 Behavioural/ Psychiatric Abnormality Thyroid stimulating hormone excess Gait apraxia Spasticity increased exploration in new environment increased dopamine level hyperactivity hyperactivity Behavioral abnormality Abnormality of the endocrine system abnormal locomotor behavior Abnormal voluntary movement Patient phenotypes Sh3kbp1 tm1Ivdi -/-
- 44. What if there aren’t any similar diseases or models? YARS MARS IARSIL41L AARSIARS2 Abnormal stereopsis Choreoathetosis Microcephaly Akinesia Visual impairment Myoclonus Microcephaly Myoclonus abnormal visual perception Involuntary movements Microcephaly musculoskeletal movement phenotype Patient phenotypes Combined Oxidative Phosphorylation Deficiency 14 FARS2 WARS2 ? AIMP1 UDP_1166 ➔ Exomiser can utilize phenotypic similarity via the interactome
- 45. Outline Semantic Diagnosis of known diseases Semantic similarity across species Combining Exome analysis with cross- species semantic phenotyping How much phenotyping is enough?
- 46. How does the clinician know they’ve provided enough phenotyping? How many annotations…? How many different categories? How many within each?
- 47. Method Create a variety of “derived” diseases that are less- specific Assess the change in similarity between the derived disease and it’s parent. Ask questions: Is the derived disease still considered similar to the original disease? …or more similar to a different disease? Is it distinguishable beyond random?
- 48. Image credit: Viljoen and Beighton, J Med Genet. 1992 Example: Schwartz-jampel Syndrome, Type I Rare disease Caused by Hspg2 mutation, a proteoglycan ~100 phenotype annotations
- 49. Example: Schwartz-jampel Syndrome, Type I to test influence of a single phenotypic category
- 50. Schwartz-jampel Syndrome derivations to test influence of a single phenotypic category
- 52. Example: Schwartz-jampel Syndrome, Type I * * * ➔When averaged over all diseases, the absence of a single phenotypic category has far less impact when there’s more breadth in annotations
- 53. How much phenotyping is enough? • How many annotations…? • How many different categories? • How many within each?
- 54. Annotation Sufficiency Score • Measurement of breadth and depth of an phenotype profile • Uses human disease, mouse and fish* gene phenotype profiles to seed the individual phenotype scores • Custom queries available via REST services • http://monarchinitiative.org/page/services *soon to add more species
- 56. Conclusions Semantic representation of patient phenotypes can aid disease diagnosis There exists a lot of phenotype data in model organisms that is complementary to known human data Ontological integration and use of cross-species inferencing can aid prioritization of variants The entire cross-species corpus can be utilized to support quality assurance processes for phenotype data capture
- 57. NIH-UDP William Bone Murat Sincan David Adams Amanda Links David Draper Joie Davis Neal Boerkoel Cyndi Tifft Bill Gahl OHSU Nicole Vasilesky Matt Brush Bryan Laraway Shahim Essaid Lawrence Berkeley Nicole Washington Suzanna Lewis Chris Mungall UCSD Amarnath Gupta Jeff Grethe Anita Bandrowski Maryann Martone U of Pitt Chuck Boromeo Jeremy Espino Becky Boes Harry Hochheiser Acknowledgments Sanger Anika Oehlrich Jules Jacobson Damian Smedley Toronto Marta Girdea Sergiu Dumitriu Heather Trang Mike Brudno JAX Cynthia Smith Charité Sebastian Kohler Sandra Doelken Sebastian Bauer Peter Robinson Funding: NIH Office of Director: 1R24OD011883 NIH-UDP: HHSN268201300036C
- 59. Candidate gene prioritization Phenot ypic inf or mat ionGenet ic inf or mat ion gene/ gene pr oduct Inf o Phenotypes collected for individual patients Sequences from an individual,family,or related group Candidate interpretation Human sequence reference sequences (e.g.reference sequence,1K genome data, genomic location) Community phenotype data (e.g. literature MODS,KOMP2,OMIM, EHRs,GWAS,ClinVar,disease specific repositories,etc.) Pathway Functional (GO) Gene expression, OMICS data Protein-Protein Interactions Enrichment analysis (e.g.GATACA,Galaxy) Combined variant + phenotype candidate reporting(e.g.Exomizer) BiomedicalKnowledgeIndividual'sInformation Phenotypic comparison methods Variant calling (e.g.GATK) Pathogenicity /Impact calling (e.g. VAAST,SIFT) Orthologs Network module analysis
- 60. Survey of Annotations in Disease Corpus* ➔Most diseases impact >1 system
- 61. PhenoViz: Integrate all human, mouse, and fish data to understand CNVs Desktop application for differential diagnostics in CNVs Explain manifestations of CNV diseases based on genes contained in CNV E.g., Supravalcular aortic stenosis in Williams syndrome can be explained by haploinsufficiency for elastin Double the number of explanations using model data Doelken, Köhler, et al. (2013) Dis Model Mech 6:358-72













![B6.Cg-Alms1foz/fox/J
increased weight,
adipose tissue volume,
glucose homeostasis altered
ALSM1(NM_015120.4)
[c.10775delC] + [-]
GENOTYPE
PHENOTYPE
obesity,
diabetes mellitus,
insulin resistance
increased food intake,
hyperglycemia,
insulin resistance
kcnj11c14/c14; insrt143/+(AB)
Models recapitulate various
phenotypic aspects of
disease
?](https://image.slidesharecdn.com/udnpresentation-140715134251-phpapp01/85/Use-of-semantic-phenotyping-to-aid-disease-diagnosis-15-320.jpg)